Abstract
Angelica sinensis is a famous traditional Chinese medicinal herb which is used with a long history in China. However, presently, it is on the verge of extinction due to over-exploitation and habitat destruction. To conduct population genomic studies and offer strategies of conservation and management, we sequenced the complete chloroplast genome of A. sinensis using the next-generation sequencing technology. The complete cp genome is 142,822 bp in length, which contains a large single-copy region (LSC) of 99,964 bp, a small single-copy (SSC) region of 17,750 bp, and two inverted repeat (IRa and IRb) regions of 12,554 bp. The G + C content of cp genome is 37.4%. The genome comprises a total of 128 genes, including 83 protein-coding genes, eight ribosomal RNA(rRNA) genes and 35 transfer RNA (tRNA). Phylogenetic inference indicated that A. sinensis was closely related to A. gigas, A. decursiva, A. nitida and A.dahurica.
Angelica sinensis (Oliv.) Diels is a traditional Chinese medicinal herb for medical use with a long history in China (Zhang et al. Citation2012). Danggui (Angelicae Sinensis Radix), the dried root of A. sinensis, is thought to enrich and activate the blood, regulate menstruation and relax bowel (National Pharmacopoeia Committee Citation2015). Danggui almost originates from single cultivated variety in China, which has performed resources decline, resistance reduction, yield and quality decline (Sun et al. Citation2009). Meanwhile, assessment on the wild A. sinensis showed that the reserves have sharply declined and it is on the verge of extinction as a result of over-exploitation and habitat destruction (Huang Citation2006). As we know, wild relatives are the natural gene pool for germplasm innovation and variety improvement of cultivated plants. A study on population genomics of wild A. sinensis with the complete chloroplast genome data might offer effective conservation and management strategies. Therefore, we sequenced the complete chloroplast genome of A. sinensis.
One individual of A. sinensis was collected from Min county, Ganshu province, China (N38°34'39" E106°13'32") and deposited in the herbarium of school of Traditional Chinese Medicine, Southern Medical University. Total genomic DNA was extracted with the modified CTAB method (Yang et al. Citation2014). About 100–200 ng high-quality DNA was used for library construction and sequencing was run on the Illumina Hiseq 4000 instrument (BGI, Shenzhen, China). The chloroplast genome of A. sinensis was reconstructed using Geneious 8.1 (Kearse et al. Citation2012) with A. decursiva (accession no. KT781591) as the reference. The chloroplast genome of A. sinensis was annotated using DOGMA (Wyman et al. Citation2004) and deposited in Genbank (Accession no.: MH430891).
The length of the complete chloroplast genome of A. sinensis is 142,822 bp, including a large single-copy (LSC) region of 99,964 bp, a small single-copy (SSC) region of 17,750 bp and two inverted repeat (IRa and IRb) regions of 12,554 bp. The G + C content of cp genome of A. sinensis is 37.4%. The cp genome comprises a total of 128 genes, including 83 protein-coding genes, eight ribosomal RNA (rRNA) genes, and 35 transfer RNA (tRNA) genes. Fifteen genes (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA) contain one intron, while two genes (clpP, ycf3) have two introns. And, a trans-splicing gene was found (rps12).
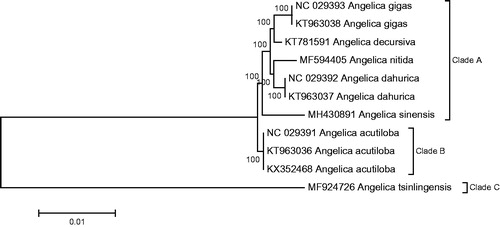
Eleven Angelica plastome (seven speices, including A. sinensis) were used to infer the inter-relationships of the genus Angelica. All sequences were aligned with MAFFT (Katoh and Standley Citation2013). Phylogenetic reconstruction was performed with MEGA5.1 based on the neighbour-joining (NJ) analysis (Tamura et al. Citation2011). Phylogenetic tree showed these seven species clustered into three clades (Clade A, Clade B and Clade C) with 100% bootstrap value. A. sinensis was closely related to A. gigas, A. decursiva, A. nitida and A. dahurica (Clade A), and A. acutiloba and A. tsinlingensis respectively clustered into Clade B and Clade C, which is consistent with the phylogenetic results that reported in study of Zhang et al. (Citation2018) ().
Disclosure statement
The authors declared they had no conflicts of interest.
Additional information
Funding
References
- Huang LQ. 2006. Molecular pharmacognosy. Bejing: Peking University Medical Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- National Pharmacopoeia Committee. 2015. Pharmacopoeia of the People’s Republic of China. Bejing: China Medical Science and Technology Press.
- Sun HM, Zhang BG, Qi YD, Zhang Z, Guo ZX. 2009. Survey and analysis on the resource of Angelica sinensis. Chin Agr Sci Bull. 25:437–441.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Res. 14:1024–1031.
- Zhang SZ, He LP, Han LM, Yang WX, Liu LL. 2012. Study on wild Angelica resources. Modern Chin Med. 14:33–36.
- Zhang H, Wang XF, Cao D, Niu JF, Wang ZZ. 2018. The complete chloroplast genome sequence of Angelica Tsinlingensis (Apioideae). Mitochondrial DNA Part B. 3:480–481.