Abstract
To provide genetic data for the cricket subfamily Eneopterinae, the complete mitochondrial genome (mitogenome) of a bush cricket Xenogryllus marmoratus (Haan, 1844) was determined. This mitogenome was 15,762 bp in size containing the typical 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a control region (CR). Compared with the ancestral insect gene arrangement, X. marmoratus had an inversion involving trnE-trnS1-trnN. Phylogenetic analysis supported that X. marmoratus was sister to a formerly sequenced species Cardiodactylus muiri of Eneopterinae. This study provides new data for further phylogenetic studies of Eneopterinae or higher taxa.
Eneopterinae Saussure, 1874 is a subfamily of insects in the cricket family Gryllidae. This subfamily has a worldwide distribution and encompasses 249 valid extant species (Cigliano et al. Citation2018). Its taxonomy and phylogeny is a research focus in recent years (Robillard and Desutter-Grandcolas Citation2008; Nattier et al. Citation2011; Vicente et al. Citation2017). Although mitogenome represents a powerful tool to elucidate insect phylogenetic relationships (Cameron Citation2014), its utilization in Eneopterinae is not feasible so far due to the availability of only one mitogenome in GenBank (Dong et al. Citation2017). To provide genetic data for further phylogenetic studies, the complete mitogenome of X. marmoratus was sequenced in this study.
Samples of X. marmoratus were collected from Shanghai, China (31.188°N, 121.437°E) and preserved in 100% ethanol at 4 °C. The voucher specimens were deposited in the Institute of Apicultural Research, Chinese Academy of Agricultural Sciences. Whole genomic DNA was extracted from a single sample using a DNeasy Blood & Tissue kit (Qiagen, Valencia, CA). High-throughput sequencing was executed on the HiSeq 2500 platform (Illumina Inc.). Assembly and annotation of the full mitogenome were performed as per Ma and Li (Citation2018). Long non-coding regions were amplified via PCR followed by Sanger sequencing to validate their sequence accuracy. Phylogeny of the family Gryllidae was reconstructed based on concatenated sequences of the 37 genes. The partitioned models selected by PartitionFinder v2.1.1 (Lanfear et al. Citation2017) were used in MrBayes v3.2 (Ronquist et al. Citation2012) for the Bayesian inference.
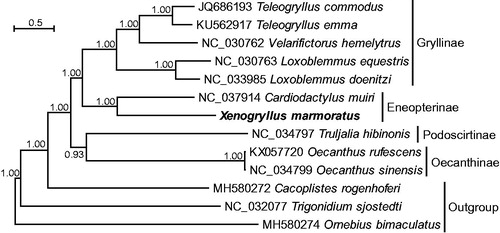
The complete mitogenome of X. marmoratus (GenBank accession: MK033622) was a circular molecule of 15,762 bp in length with a strong A + T-bias in nucleotide composition (40.8% A, 31.3% T, 19.2% C, and 8.7% G). Like other insect mitogenomes, it contained a typical set of 37 genes, i.e. two rRNA genes, 22 tRNA genes, and 13 PCGs. ATN acted as start codons of all PCGs except cox1 starting with TCG, which has been proposed to be a start codon of cox1 based on transcript information (Stewart and Beckenbach Citation2009; Ma and Li Citation2018). X. marmoratus possessed an inversion of the ancestral trnN-trnS1-trnE to trnE-trnS1-trnN, a common rearrangement pattern found in all sequenced mitogenomes of the family Gryllidae as well as closely related families, Phalangopsidae and Trigonidiidae (Ma and Li Citation2018). In addition, X. marmoratus contained long non-coding regions with the CR being the largest one (711 bp), followed by 174 bp between trnH and nad4, 99 bp between trnS2 and nad1, and 94 bp between trnQ and trnM. The phylogenetic tree showed that the newly sequenced X. marmoratus was sister to a formerly sequenced C. muiri (Eneopterinae) with a high posterior probability (). Sequencing the mitogenome of X. marmoratus provides new data for further phylogenetic studies involving the subfamily Eneopterinae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Cigliano MM, Braun H, Eades DC, Otte D. 2018. Orthoptera Species File. Version 5.0/5.0 http://Orthoptera.SpeciesFile.org (accessed 11 October 2018).
- Dong JJ, Vicente N, Chintauan-Marquier IC, Ramadi C, Dettai A, Robillard T. 2017. Complete mitochondrial genome and taxonomic revision of Cardiodactylus muiri Otte, 2007 (Gryllidae: Eneopterinae: Lebinthini). Zootaxa. 4268:101–116.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Ma C, Li J. 2018. Comparative analysis of mitochondrial genomes of the superfamily Grylloidea (Insecta, Orthoptera) reveals phylogenetic distribution of gene rearrangements. Int J Biol Macromol. 120:1048–1054.
- Nattier R, Robillard T, Desutter-Grandcolas L, Couloux A, Grandcolas P. 2011. Older than New Caledonia emergence? A molecular phylogenetic study of the eneopterine crickets (Orthoptera: Grylloidea). J Biogeogr. 38:2195–2209.
- Robillard T, Desutter-Grandcolas L. 2008. Clarification of the taxonomy of extant crickets of the subfamily Eneopterinae (Orthoptera: Grylloidea; Gryllidae). Zootaxa. 1789:66–68.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stewart JB, Beckenbach AT. 2009. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene. 445:49–57.
- Vicente N, Kergoat GJ, Dong JJ, Yotoko K, Legendre F, Nattier R, Robillard T. 2017. In and out of the Neotropics: historical biogeography of Eneopterinae crickets. J Biogeogr. 44:2199–2210.