Abstract
Pseudostellaria okamotoi Ohwi is the endemic species with limited habitat which is subalpine area in central Southern part of the Korean peninsula. In this study, we presented the complete chloroplast genome of P. okamotoi of which length is 149,653 bp. It consists of four subregions: 81,291 bp of large single copy (LSC) and 16,958 bp of small single copy (SSC) regions are separated by 25,702 bp of inverted repeat (IR) regions. It includes 126 genes (81 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.5% and those in the LSC, SSC, and IR regions are 34.3%, 29.3%, and 42.4%, respectively.
Pseudostellaria okamotoi Ohwi is an endemic species of Korea which was reported as a new species in 1936 (Ohwi Citation1936). It is clearly distinguished from other Pseudostellaria species by having a line on a long pedicel. It is only distributed in Mt. Deogyu and Mt. Jiri in the central Southern part of Korean peninsula included in sub-alpine area (Park Citation2007; Shin et al. Citation2010). The distribution of alpine plants is gradually decreasing due to the increase in temperature due to climate change (Walther et al. Citation2005; Dullinger et al. Citation2012), so that P. okamotoi distributed in sub-alpine areas should be affected by climate changes which will cause extinction. Even though it is Korean endemic species and threated species by recent climate changes, little research has been conducted. As a first step to acquire genetic information of P. okamotoi for its conservation as well as for determining its phylogenetic position with Pseudostellaria longipedicellata of which chloroplast genome has recently been sequenced (Kim et al. Citation2018), we completed chloroplast genome of P. okamotoi.
Pseudostellaria okamotoi was collected in Mt. Deogyu, Jeollabuk province, Korea (Voucher in InfoBoss Cyber Herbarium (IN); Y. Kim, IB-00655). Total genomic DNA was extracted from fresh leaves of P. okamotoi by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011) under the in-house bioinformatic pipeline (the Plant Chloroplast Database; in preparation). Geneious R11 11.1.5 (Biomatters Ltd., Auckland, New Zealand) was used for genome annotation.
The chloroplast genome of P. okamotoi (MH879018) is 149,653 bp and composed of four subregions: 81,291 bp of large single copy (LSC), 16,958 bp of small single copy (SSC) regions, and 25,702 bp of a pair of inverted repeats (IRs). It contains 126 genes (81 protein-coding genes, eight rRNAs, and 37 tRNAs); 18 genes (seven protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in inverted repeat regions. The overall GC content of P. okamotoi is 36.5% and those in the LSC, SSC, and IR regions are 34.3%, 29.3%, and 42.4%, respectively.
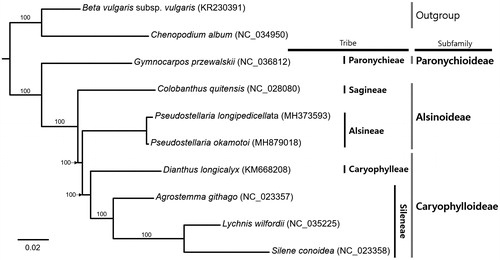
Seven complete chloroplast genomes of Caryophyllaceae and two species from Amaranthaceae as outgroup were used for constructing maximum likelihood method phylogenetic tree. Sequence alignment was conducted by MAFFT 7.388 and phylogenetic tree was constructed by IQ-TREE 1.6.6 (Katoh and Standley Citation2013; Nguyen et al. Citation2015) with 1,000 replicate bootstrap analyses. The phylogenetic tree shows that P. okamotoi is sister to P. longipedicellata forming monophyletic group closely related with Caryophylloideae (). This chloroplast genome will be a useful resource for an understanding phylogenetic relationship as well as for conservation of this species under recent global climate changes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dullinger S, Gattringer A, Thuiller W, Moser D, Zimmermann NE, Guisan A, Willner W, Plutzar C, Leitner M, Mang T, et al. 2012. Extinction debt of high-mountain plants under twenty-first-century climate change. Nat Clim Change. 2:619.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Katoh Kim Y, Heo KI, Lee S, Park J. 2018. Complete chloroplast genome sequence of the Pseudostellaria longipedicellata S. Lee, K. Heo &Kim (Caryophyllaceae). Mitochondrial DNA Part B. 3(2):1296–1297.
- Kim Y, Heo K I, Lee S,Park J. 2018. Complete chloroplast genome sequence of the Pseudostellaria longipedicellata S. Lee, K. Heo & SC Kim (Caryophyllaceae). Mitochondrial DNA Part B. 3(2):1296–1297.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ohwi J. 1936. Plantae novae Japonicae (II). J Jpn Bot. 12:379–390.
- Park C. 2007. The genera of vascular plants of Korea. Seoul: Academic Publishing 1482p ISBN.
- Shin H-T, Yi M-H, Yoon J-W, Yoo J-H, Lee B-C, Park E-H. 2010. Distribution of rare plants and endemic plants in Jirisan national park. J Asia-Pac Biodivers. 3:219–222.
- Walther GR, Beißner S, Burga CA. 2005. Trends in the upward shift of alpine plants. J Veg Sci. 16:541–548.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.