Abstract
Rehmannia glutinosa is a plant used as traditional medicine for its various tonic effects in Korea and China. In this study, chloroplast genomes of two R. glutinosa were completed by de novo assembly using whole-genome Illumina sequence data. The length of chloroplast genomes of R. glutinosa collected from China and Korea was 153,680 bp and 153,499 bp, respectively. A total of 114 coding regions were predicted in both R. glutinosa including 80 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. We identified abundant intra-species diversity of 87 InDels and 147 SNPs, among three R. glutinosa chloroplast genome sequences including one from GenBank. The phylogenetic analysis showed that these R. glutinosa were closely clustered with related Rehmannia species, separated from other Orobanchaceae and Scrophulariaceae species.
Rehmannia glutinosa belongs to the Orobanchaceae family. The roots of R. glutinosa have been used as ingredients for traditional medicines in Korea and China (Kim et al. Citation2008; Zhang et al. Citation2008). The R. glutinosa is effective in haemostasis, anti-tumour, anti-aging, and anti-ulcer in stomach. It has been cultivated mainly in the western part of China but is also distributed in wild habitats such as mountain slopes and trails, ranging from near sea level to 1100 m (Zhang et al. Citation2008). Chloroplast genome sequences provide good genomic tools for studying genetic diversity and authentication of the plant (Joh et al. Citation2017; Kim et al. Citation2017; Nguyen et al. Citation2017; Nguyen et al. Citation2018). We characterized the chloroplast genome sequences of R. glutinosa to contribute to genomic studies of R. glutinosa.
Wild R. glutinosa plant was provided by the Institute of Medicinal Plant Development (IMLAD) in China (R. glutinosa_Chinese), and a cultivating variety was provided by the Rural Development Administration (RDA) in Korea (R. glutinosa_‘Jiwhang1’), respectively. DNA was extracted from leaf tissues using a modified cetyltrimethylammonium bromide (CTAB) method (Allen et al. Citation2006) and sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA). A total of 1.6 and 1.5 Gbp of paired-end reads were obtained from R. glutinosa_Chinese and R. glutinosa_‘Jiwhang1,’ respectively. They were assembled with the de novo assembly method using CLC genome assembler, ver 4.6 (Qiagen, Aarhus, Denmark), following the previous research (Kim et al. Citation2015a, Citation2015b) with the chloroplast genome sequence of Scrophularia takesimensis as a reference (KM590983). Gene annotation was conducted using GeSeq (Tillich et al. Citation2017) and manually curated using BLAST searches.
The complete chloroplast genomes of R. glutinosa_Chinese and R. glutinosa_‘Jiwhang1’ are 153,690 and 153,499 bp in total length, respectively, with circular structures. The genome was sorted into four sections as one large single copy (LSC) region of 84,590–84,401 bp, one small single copy (SSC) region of 17,592–17,600 bp, and a pair of inverted repeat (IR) regions of 25,749 bp. A total of 114 coding regions were predicted including 80 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. The complete chloroplast genome sequences were submitted to GenBank (Accession numbers: MG977438, MG977439). Comparative analysis of three chloroplast genomes including an available sequence in GenBank (NC034308) was conducted to observe intraspecific diversity and a total of 147 SNPs and 87 InDels were identified.
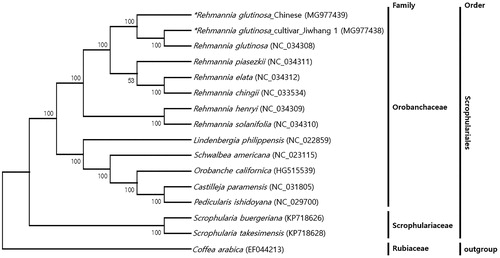
The phylogenetic relationship of two R. glutinosa plastid genomes was constructed with closely related plants belonging to the Scrophulariales family, including the R. glutinosa previously registered in GenBank, 10 species in the Orobanchaceae family, 2 species in the Scrophulariaceae family, and Coffea arabica as an outgroup. Multiple alignments were carried out by MAFFT based on entire chloroplast genome sequences (Katoh et al. Citation2009). A phylogenetic tree was produced with the neighbour-joining method in MEGA 6.0 using 1000 bootstrap replicates (Tamura et al. Citation2013). The phylogenetic tree clearly classified 13 Orobanchaceae species and 2 Scrophulariaceae species. In the Orobanchaceae group, eight Rehmannia species were grouped into a clade and the other five genera were clustered together ().
Figure 1. Phylogenetic tree of 14 species including R. glutinosa based on the entire chloroplast genome sequences. A phylogenetic tree was produced with the neighbour-joining method in MEGA 6.0 using 1000 bootstrap replicates. The number in the nodes is bootstrap values from 1000 replicates. Coffea arabica (EF044213) was used as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320–2325.
- Joh HJ, Kim N-H, Jayakodi M, et al. 2017. Authentication of golden-berry P. ginseng Cultivar ‘Gumpoong’ from a landrace ‘Hwangsook’ based on pooling method using chloroplast-derived markers. Plant Breed Biotechnol. 5:16–24.
- Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 537:39–64.
- Kim D, Park C, Park H, Park C, Sung J, Yu H, Kim G, Seong N, Kim J, Kim M. 2008. A new high-quality, disease resistance and high-yielding Rehmannia glutinosa cultivar," Kokang". Korean J Breed Sci. 40:84–87.
- Kim K, Lee S-C, Lee J, Lee HO, Joh HJ, Kim N-H, Park H-S, Yang T-J. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PloS One. 10:e0117159.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kim K, Nguyen VB, Dong J, Wang Y, Park JY, Lee S-C, Yang T-J. 2017. Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species. Sci Rep. 7:4917.
- Nguyen VB, Park H-S, Lee S-C, Lee J, Park JY, Yang T-J. 2017. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J Agric Food Chem. 65:6298–6306.
- Nguyen VB, Giang VNL, Waminal NE, Park H-S, Kim N-H, Jang W, Lee J, Yang T-J. 2018. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique SNP markers. J Ginseng Res. (In Press).
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Zhang R-X, Li M-X, Jia Z-P. 2008. Rehmannia glutinosa: review of botany, chemistry and pharmacology. J Ethnopharmacol. 117:199–214.
