Abstract
The nearly complete mitochondrial genomes of four bumblebee species (Bombus sibiricus, Bombus asiaticus, Bombus pyrosoma, and Bombus kashmirensis) were sequenced. Each of the four mitogenomes contained 38 sequence elements including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and an AT-rich control region (D-loop). Most elements showed a consistent order among the four mitogenomes, except for two tRNA genes (tRNA-Ala and tRNA-Met). Phylogenetic analyses based on the concatenated sequence of the 13 PCGs showed that Alpigenobombus and Sibiricobombus had the closest relationship, while Melanobombus showed a more basal position.
Bumblebees are a group of creatures belonging to the genus Bombus (Hymenoptera: Apidae). There are about 250 known bumblebee species subdivided into 15 subgenera, mainly limited to temperate, alpine and arctic zones of the Northern Hemisphere (Williams et al. Citation2008). Alpigenobombus, Melanobombus, and Sibiricobombus were three sibling subgenera within Bombus. Many species from these subgenera were widespread pollinators in Northern Asia (An et al. Citation2014). However, the phylogenetic relationships among these subgenera were not well clarified (Cameron et al. Citation2007). Here we present four mitochondrial genomes (B. sibiricus, B. asiaticus, B. pyrosoma, and B. kashmirensis) and combine with other bumblebee mitogenomes available in the GenBank, we tested the phylogenetic relationships among the three subgenera.
Samples of the four species were collected from Xining, China (N36°34′, E101°45′). Specimens were stored in the Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, China, accession number: XN-2017-001∼XN-2017-004. Illumina sequencing was performed following the manufacturer’s instructions. The paired-end 500 bp libraries were sequenced on a HiSeq2000 platform with both directions of 150 bp reads. The reads were used for de novo assembly with MEGAHIT v1.1.2 (Li et al. Citation2016). The assembled mitogenomes were annotated using the MITOS web server (Bernt et al. Citation2013). The protein-coding genes (PCGs) of each mitogenome were concatenated for phylogenetic analysis using MrBayes v3.2.6 (Huelsenbeck and Ronquist Citation2001). The best-fit nucleotide substitution model (GTR + I + G) for tree reconstruction was determined by the Akaike Information Criterion implemented in jModelTest v2.1.6 (Darriba et al. Citation2012).
The mitogenomes of the four species were nearly complete with 20,048 bp, 19,752 bp, 18,897 bp, and 16,793 bp respectively for B. sibiricus, B. asiaticus, B. pyrosoma, and B. kashmirensis. They were deposited to GenBank with accession number MH998258∼ MH998261. Each mitogenome contained 13 PCGs, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and an AT-rich control region (D-loop), similarly as in other Bombus mitogenomes (Zhao et al. Citation2017; Takahashi et al. Citation2018). Most elements showed a consistent order among the four mitogenomes, except for two tRNA genes: B. sibiricus, B. pyrosoma, and B. kashmirensis showed a tRNA-Ala to tRNA-Met order (from 5′ to 3′), while B. asiaticus had a reversed direction.
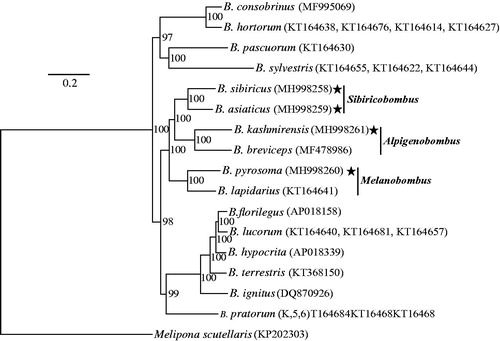
The generated Bayesian tree showed accurate interspecific phylogenetic relationships with high (> 95%) bootstrap values in the nodes (). Of the three subgenera emphasized in this study, Alpigenobombus and Sibiricobombus had the closest relationship, while Melanobombus showed a more basal position. Although all bumblebee species in this study had also been analyzed using several genes in a previous study (Cameron et al. Citation2007), the phylogenetic relationship among the three subgenera was not well solved (with low bootstrap values), probably because of not sufficient polymorphism information. Our results, with 100% bootstrapping rates on the related nodes, emphasized the value of mitogenomes on phylogenetic analyses.
Figure 1. Phylogenetic analyses of bumblebee mitochondrial genomes with Melipona scutellaris as an outgroup. The stars indicate the focal mitochondrial genomes in this study. Numbers beside each node represent percentages of Bayesian bootstrap values. Species names are followed by the GenBank accession numbers of their mitochondrial genomes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- An J, Huang J, Shao Y, Zhang S, Wang B, Liu X, Wu J, Williams PH. 2014. The bumblebees of North China (Apidae, Bombus Latreille). Zootaxa. 3830:1–89.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Cameron SA, Hines HM, Williams PH. 2007. A comprehensive phylogeny of the bumble bees (Bombus). Biol J Linn Soc. 91:161–188.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. 2016. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 102:3–11.
- Takahashi JI, Sasaki T, Nishimoto M, Okuyama H, Nomura T. 2018. Characterization of the complete sequence analysis of mitochondrial DNA of Japanese rare bumblebee species Bombus cryptarum florilegus. Conservation Genet Resour. 10:387–391.
- Williams PH, Cameron SA, Hines HM, Cederberg B, Rasmont P. 2008. A simplified subgeneric classification of the bumblebees (genus Bombus). Apidologie. 39:46–74.
- Zhao X, Huang J, Sun C, An J. 2017. Complete mitochondrial genome of Bombus consobrinus (Hymenoptera: Apidae). Mitochondrial DNA Part B. 2:770–772.
