Abstract
Oncodostigma hainanense is an endangered species with extremely small populations due to human activities in China. This study presents the chloroplast genome of the plant by de novo assembly using the whole-genome sequencing data. The size of the complete chloroplast genome was 160,497 bp in length, comprising a large single copy region (LSC) of 89,424 bp, a small single copy region (SSC) 18,948 bp, and inverted repeat regions (IRS) of 26,061 bp. A total of 135 genes, including 85 protein-coding genes, 42 tRNA genes and eight rRNA genes, were predicted from the chloroplast genomes. Among them, 17 genes occur in IRS, containing six protein-coding genes, seven tRNA genes and four rRNA genes. The GC content of O. hainanense was 35.59%. Phylogenetic analysis with seven other species revealed that O. hainanense was clustered with Annona cherimola.
Oncodostigma hainanense (Merrill) Tsiang & P. T. Li (Annonaceae) is an evergreen tree with a height to 15 m. It is distributed in parts of Hainan and Guangxi province in China, inhabiting dense forest along valleys. Oncodostigma hainanense is an endangered species and its population has been drastically reduced due to human activities (Shao and Xu Citation2015). Early studies of O. hainanense have mainly focused on the leaf morphology (Sun et al. Citation2002), biochemical activity (Wang Citation2010) and pollen morphology of the plant (Liu and Xu Citation2012). Besides, Shao and Xu (Citation2015) studied the development and function of the strumae of O. hainanense, which was found to provide flower visitors a nutritious and safe place for mating, oviposition, brooding and perhaps some food such as polysaccharides. Complete chloroplast sequences can provide insights into the evolutionary history and natural selection process for many plants (Inoue Citation2011; Zheng et al. Citation2016). In this study, we reported and characterized the complete chloroplast genome sequence of O. hainanense to contribute to further phylogenetical and protective studies of this plant.
The plant samples were collected in Qionghai county, Hainan Island (N19°10′10.11ʺ, E110°23′22.54ʺ) China. The specimen was deposited at the botany laboratory of Hainan Normal University, Haikou, China. Total DNA was isolated from fresh leaves of five individuals of O. hainanense, by using a Dneasy Plant Mini kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. Genome sequencing was performed on an Illumina Hiseq X Ten platform (Illumina, San Diego, CA) with paired-end reads of 150 bp. In total, 10.2 Mb short sequence data with Q20 was 97.57% were obtained. Low-quality reads were filtered out and the remaining high-quality reads were used to assemble the chloroplast genome in SOAPdenovo (Luo et al. Citation2012). The genes in the chloroplast genome were annotated step by step using the DOGMA program (Wyman et al. Citation2004). The complete chloroplast genome was submitted to GenBank under the accession numbers of MK035708.
The size of O. hainanense complete chloroplast genome was 160,497 bp in length, comprising a large single copy region (LSC) of 89,424 bp, a small single copy region (SSC) 18,948 bp, and with a pair of repeat regions (IRS) of 26,061 bp. It has a GC content of 35.59% and contains of 135 genes, including 85 protein-coding genes, 42 tRNA genes and eight rRNA genes. Among the protein-coding genes, two genes (rps12 and ycf3) were found to have three introns while other ten genes (rps16, rpl2, rpl16, rpl22, atpF, rpoC1, clpP, petB, petD and ndhB) have two introns.
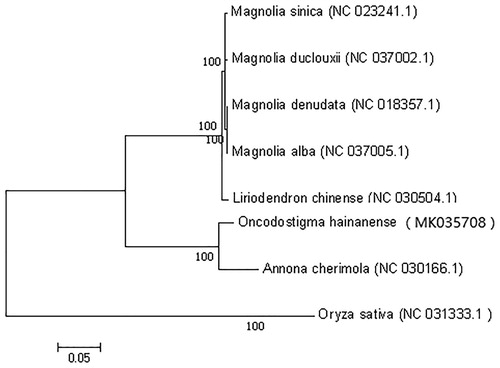
The phylogenetic relationship of O. hainanense was studied with seven other previously reported complete chloroplast genomes were downloaded for analysis. Maximum-likelihood (ML) phylogenetic tree of eight species with Oryza sativa as an outgroup () by using the maximum-likelihood algorithm was conducted with RaxML (Stamatakis Citation2014) implemented in Geneious ver. 10.1 (Kearse et al. Citation2012). Our study provides essential and important DNA molecular data for phylogenetic and evolutionary analysis for O. hainanense.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Inoue K. 2011. Emerging roles of the chloroplast outer envelope membrane. Trends Plant Sci. 16:550–557.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu Y, Xu F. 2012. Pollen morphology of four selected species in the Annonaceae. Plant Diversity Resour. 34:443–452.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Shao YY, Xu FX. 2015. Development and function of the strumae on the adaxial sides of the inner petals of Oncodostigma hainanense. Plant Sci J. 33:595–601.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sun T, Chen XF, Yang BY, Wu H, Li BT. 2002. Studies on epidermal micromorphology of eight genera in Annonaceae from China by SEM. J Chinese Electron Microsc Soc. 21:146–152.
- Wang T. Effects of traditional Chinese medicinal herbal extracts on HIV-1 replication. 2010. Faculty of the University Graduate School in partial fulfillment of the requirements for the degree Master of Science in the Department of Microbiology and Immunology, Indiana University.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zheng H, Fan L, Wang T, Zhang L, Ma T, Mao K. 2016. The complete chloroplast genome of populus rotundifolia, (salicaceae.). Conserv Genetics Resour. 8:1–3.