Abstract
Yunnanilus jinxiensis is an endemic cave-dwelling loach that only lives in the karst cave of Jingxi and Tiandong county, Southwestern China. The complete mitochondrial genome is 16,561 bp in length, consisting of 37 genes coding for 13 proteins, two rRNAs, 22 tRNAs, and one control region. Phylogenetic analysis using mitochondrial genomes of 14 species showed that Yunnanilus is mainly related to the genus Orenectes.
Yunnanilus jinxiensis, an endemic cave-dwelling loach, is first found in the karst cave of Jingxi county, Southwestern China (Zhu et al. Citation2009). Till date, there are 32 effective species in this genus Yunnanilus, and only five of them distribute in Guangxi Province (Chen 2013; Jiang et al. Citation2016). This species is larger than most of the another Yunnanilus species in size, and it usually migrate outside the cave for reproduction in June or July each year. In this study, we first determined the complete mitochondrial genome of Y. jinxiensis, and analyzed the phylogenetic relationship of the loach in Nemacheilidae fishes, contributing to aid further genetic and evolutionary researches of this fish species.
The sample of Y. jinxiensis was collected from the underground river located in Tiandong County of Guangxi, China (23°32'28.33"N, 107 11'11.62"E), and its muscle samples were fixed in 95% ethanol and preserved in Liuzhou Aquaculture Technology Extending Station, Liuzhou, China. Total genomic DNA was extracted from the fins through using traditional phenol-chloroform extraction method (Taggart et al. Citation1992). Then, we have sequenced the complete mitochondrial genome of Y. jinxiensis using the Illumina Hiseq4000 platform with de novo strategy (Tang et al. Citation2015), and submitted the genome into GenBank with accession number MG818720.
The total length of the complete mitochondrial DNA of Y. jinxiensis was 16,561 bp, and its nucleotide base composition was 31.00% A, 27.58% C, 16.02% G, 25.40% T, and an AT bias of 56.40%. This mitogenome also had a typical structure and gene order as most vertebrates, consisted of 13 protein-coding (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes and one A + T-rich control region (D-loop). All genes were encoded on the heavy strand but ND6 gene and eight tRNA genes. As in many Nemacheilidae fish mitogenomes, the typical ATG initiation codon appeared in all the coding genes except in COI gene, having GTG as the start codon. Meanwhile, the stop codons of the 13 PCGs had three types (TAA, TA-, and T-). The length of 22 tRNAs ranged from 66 bp (tRNACys) to 76 bp (tRNALys) in size, and the 12S and 16S rRNA genes were 956 bp and 1675 bp in length, respectively. The D-loop was located between tRNAPhe and tRNAPro with 911 bp long.
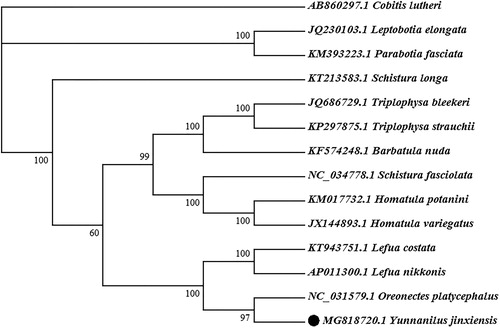
To validate the phylogenetic position of Y. jinxiensis, a neighbour-joining (NJ) tree inferred from the whole mitogenome of 13 Nemacheilidae species and one Cobitidae species (outgroup) was constructed using MEGA 7.0. Phylogenetic analysis showed that Y. jinxiensis was first clustered together with Oreonectes platycephalus, which did not belong to the genus Yunnanilus, it suggested that the genus Yunnanilus is most relate to the genus Orenectes in Nemacheilidae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen XY. 2013. Checklist of fishes of Yunnan. Zool Res. 34:281–343.
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng GM, et al. 2016. Red list of China’s vertebrates. Biodivers Sci. 24:500–551.
- Taggart J, Hynes R, Prod Uhl P, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tang M, Hardman CJ, Ji Y, Meng G, Liu S, Tan M, Yang S, Moss ED, Wang J, Yang C, et al. 2015. High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol Evol. 6:1034–1043.
- Zhu Y, Du LN, Chen XY, Yang JX. 2009. A new Nemacheiline loach of genus Yunnanilus (Balitoridae) from Guangxi, China-Yunnanilus jinxiensis. Zool Res. 30:195–198.