Abstract
The snapping shrimp genus Alpheus is the most diverse and abundant genus of Caridea. However, the phylogenetic and taxonomic studies have so far been limited. In this study, we report the third complete mitochondrial genome of Alpheus from A. hoplocheles. The mitogenome has 15,735 base pairs (60.2% A + T content) and made up of a total of 37 genes (13 protein-coding, 22 transfer RNAs and 2 ribosomal RNAs), and a control region. This study was the third available complete mitogenomes of Alpheidae and will provide useful genetic information for future phylogenetic and taxonomic classification of Alpheidae.
Caridean shrimp (Decapoda: Caridea) is one of the largest infraorders within Decapoda, which are found widely around the world in both marine and fresh waters(Williams et al. Citation2001). Alpheus is the most diverse and abundant genus of all caridean shrimp, in which 250 species and 7 groups have been revised(Shen et al. Citation2012). Alpheus inhabits marine, shallow tropical and subtropical waters, especially in Indo-West Pacific region, in which most species are important benthic crustaceans and play important roles in the marine ecosystems. However, in spite of its economic and ecological importance, there are still numerous cryptic taxa and most of which are yet undescribed(Nomura and Anker Citation2005). Moreover, adequate genetic information about the genus is still limited. Here, we report the complete mitochondrial genome sequence of Alpheus hoplocheles, which will provide a better insight into phylogenetic assessment and evolutionary classification.
Tissue samples of A. hoplocheles from five individuals were collected from GuangXi province, China (Beihai, 21.701569 N, 108.597914 E), and the whole body specimen (#GQ0403) were deposited at Marine biological Herbarium, Guangxi Institute of Oceanology, Beihai, China. The total genomic DNA was extracted from the muscle of the specimens using an SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. DNA libraries (350 bp insert) were constructed with the TruSeq NanoTM kit (Illumina, San Diego, CA) and were sequenced (2 × 150 bp paired-end) using HiSeq platform at Novogene Company, China. Mitogenome assembly was performed by MITObim(Hahn et al. Citation2013). The complete mitogenome of A. distinguendus (GenBank accession number: NC_014883) was chosen as the initial reference sequence for MITObim assembly. Gene annotation was performed by MITOS(Bernt et al. Citation2013).
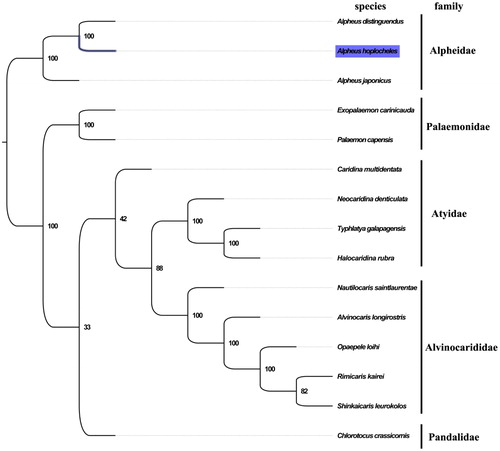
The complete mitogenome of A. hoplocheles was 15,735 bp in length (GenBank accession number: MG840649), and containing the typical set of 13 protein-coding, 22 tRNA and 2 rRNA genes, and a putative control region. The overall base composition of the mitogenome was estimated to be A 33.1%, T 27.5%, C 25.6%, and G 13.8%, with a high A + T content of 60.6%, which is similar, but slightly higher than Alpheus distinguendus (60.2%)(Qian et al. Citation2011). The result of phylogenetic tree of 15 species (including other 14 species from infraorder Caridea in NCBI) indicated the close relationship between Alpheidae and Palaemonidae at the family level (), which is consistent with the phylogenetic analyses of seven caridean shrimps based on the mitochondrial genomes(Shen et al. Citation2012). As more mitochondrial data from Alpheidae become available, phylogenetic relationships within this group will be better defined. The complete mitochondrial genome sequence of A. hoplocheles was the third sequenced mitogenome in Alpheidae, which will contribute to further phylogenetic and comparative mitogenome studies of Alpheidae, and related families.
Figure 1. Phylogenetic tree of 15 species in infraorder Caridea. The complete mitogenome is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 100 bootstrap replicates. The bootstrap values were labelled at each branch nodes. The gene's accession number for tree construction is listed as follows: Alpheus japonicus (NC_038116), Alpheus distinguendus (NC_014883), Palaemon capensis (NC_039373), Exopalaemon carinicauda (NC_012566), Caridina multidentata (NC_038067), Neocaridina denticulata (NC_023823), Typhlatya galapagensis (NC_035402), Halocaridina rubra (NC_008413), Nautilocaris saintlaurentae (NC_021971), Alvinocaris longirostris (NC_020313), Opaepele loihi (NC_020311), Rimicaris kairei (NC_020310), Shinkaicaris leurokolos (NC_037487), and Chlorotocus crassicornis (NC_035828).
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogen Evol. 69:313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Nomura K, Anker A. 2005. The taxonomic identity of Alpheus gracilipes Stimpson, 1860 (Decapoda: Caridea: Alpheidae), with description of five new cryptic species, from Japan. Crustacean Res. 34:104–139.
- Qian G, Zhao Q, Wang AN, Zhu LIN, Zhou K, Sun H. 2011. Two new decapod (Crustacea, Malacostraca) complete mitochondrial genomes: bearings on the phylogenetic relationships within the Decapoda. Zoo J Linnean Soc. 162:471–481.
- Shen X, Li X, Sha Z, Yan B, Xu Q. 2012. Complete mitochondrial genome of the Japanese snapping shrimp Alpheus japonicus (Crustacea: Decapoda: Caridea): gene rearrangement and phylogeny within Caridea. Sci China Life Sci. 55:591–598.
- Williams ST, Knowlton N, Weigt LA, Jara JA. 2001. Evidence for three major clades within the snapping shrimp genus Alpheus inferred from nuclear and mitochondrial gene sequence data. Mol Phylogen Evol. 20:375–389.
