Abstract
Paecilomyces penicillatus is the causative pathogens of white mould on cultivated Morchella. Here, we report the complete mitochondrial genome of P. penicillatus for the first time. The mitogenome is composed of circular DNA molecules, with the total length of 27,480 bp, which encoded 17 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNA), and 24 transfer RNA (tRNA) genes. The base composition of the P. penicillatus mitogenome is as follows: A (36.69%), T (35.79%), G (15.27%), and C (12.25%). Phylogenetic analysis revealed that P. penicillatus had a close relationship with Epichloe festucae and Epichloe typhina from Clavicipitaceae. This study provided important information on the evolution and taxonomy of P. penicillatus.
The genus Paecilomyces was established by Bainier, which was considered to be similar to Penicillium (Bainier Citation1907). Luangsa-Ard et al. (Citation2004) demonstrated that Paecilomyces was polyphyletic across two subclasses (Sordariomycetidae and Eurotiomycetidae) through phylogenetic analysis of the 18S rRNA. Inglis and Tigano (Citation2006) further supported the polyphyly of the genus through phylogenetic analysis of the 5.8S rRNA and internal transcribed spacer (ITS1 and ITS2) sequences from some Paecilomyces species. Paecilomyces penicillatus was originally isolated from rotten mushrooms (Samson Citation1974). He et al. (Citation2017) demonstrated that P. penicillatus was the causative pathogen of morels which produced white mold-like symptoms on the caps and stripes of Morchella. Furthermore, P. penicillatus was not monophyletic, which had an uncertain affinity with the Clavicipitaceae within the Hypocreales (Luangsa-Ard et al. Citation2005). The mitogenome of P. penicillatus reported here would provide important information on the evolution and taxonomy of P. penicillatus.
The strain was isolated from the diseased fruiting body of cultivated morels from Chengdu, Sichuan Province, China (103.87 E; 30.68 N). Koch’s postulates, morphological features, and ITS sequences analysis showed that the strain was identical to the causative agent of white mould on cultivated Morchella (He et al. Citation2017). The specimen (P. penicillatus) was stored in Sichuan Academy of Agricultural Sciences (No. SAAS_ppe1). Total DNA was extracted from the mycelia using the fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA) according to the manufacturer’s instructions. Purified DNA was used to construct the sequencing libraries following the instructions of NEBNext Ultra II DNA Library Prep Kit (NEB, Beijing, China). Mitochondrial (mt) genome sequencing was performed using an Illumina HiSeq 2500 Platform (Illumina, San Diego, CA). We performed quality control and de novo assembly of the mitogenome according to Bi (Citation2017). The SPAdes 3.9.0 (Bankevich et al. Citation2012) was used to de novo assemble the mitogenome; the MITObim 1.9 (Hahn et al. Citation2013) was used to fill the gaps between contigs. The MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and MITOS (Bernt M et al. Citation2013) tool were used for mt genome annotation of P. penicillatus. The tRNAscan-SE 2.0 (Lowe and Chan Citation2016) was used to predict tRNA genes.
The mitogenome of P. penicillatus was assembled as circular DNA molecules with 27,480 bp in length, which encoded 17 protein-coding genes (PCGs), 24 tRNA genes, and 2 ribosomal RNA genes (rnl and rns). The overall base composition of P. penicillatus is as follows: A (36.69%), T (35.79%), G (15.27%), and C (12.25%). The mitogenome of P. penicillatus was submitted to GenBank database under Accession No. MK069583.
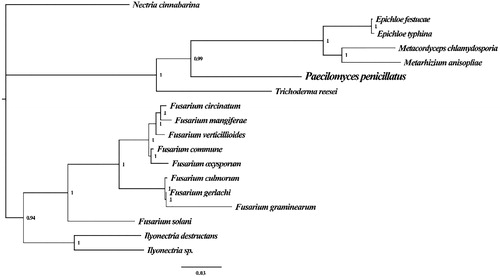
The Bayesian inference (BI) methods using the MrBayes version 3.2.6 (Ronquist et al. Citation2012) software were performed for phylogenetic analysis of P. penicillatus and 17 closely related species. The 15 typical PCGs (14 conserved core PCGs and rps3) were used to construct the phylogenetic trees, and the node support was calculated according to Bayesian posterior probabilities (BPP) (). The phylogenetic analysis revealed that P. penicillatus has a close relationship with Epichloe festucae and Epichloe typhina from Clavicipitaceae.
Figure 1. Molecular phylogenies of 18 species based on Bayesian inference analysis of the combined mitochondrial gene set (15 core protein-coding genes). Node support values are Bayesian posterior probabilities (BPP). Mitogenome accession numbers used in this phylogeny analysis: Epichloe festucae (NC_032064), Epichloe typhina (NC_032063), Metacordyceps chlamydosporia (NC_022835), Metarhizium anisopliae (NC_008068), Trichoderma reesei (NC_003388), Fusarium circinatum (NC_022681), Fusarium mangiferae (NC_029194), Fusarium verticillioides (NC_016687), Fusarium commune (NC_036106), Fusarium oxysporum (NC_017930), Fusarium culmorum (NC_026993), Fusarium gerlachii (NC_025928), Fusarium graminearum (NC_009493), Fusarium solani (NC_016680), Ilyonectria destructans (NC_030340), Ilyonectria sp. (MF924828), Nectria cinnabarina (NC_030252).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bainier G. 1907. Mycothe`que de l'e´cole de Pharmacie. XI Paecilomyces, genre nouveau de Muce´dine´ es. Bull Soc Mycol Fr. 23:26–27.
- Bi GQ. 2017. The complete mitochondrial genome of northern grasshopper mouse (Onychomys leucogaster). Mitochondrial DNA Part B. 2:393–394.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. Mitos: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- He XL, Peng WH, Miao RY, Tang J, Chen Y, et al. 2017. White mold on cultivated morels caused by Paecilomyces penicillatus. FEMS Microbiol Lett. 364:1–5.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Inglis PW, Tigano MS. 2006. Identification and taxonomy of some entomopathogenic Paecilomyces spp. (Ascomycota) isolates using rDNA-ITS Sequences. Genet Mol Biol. 29:132–136.
- Luangsa-Ard JJ, Hywel-Jones NL, Samson RA. 2004. The polyphyletic nature of Paecilomyces sensu lato as revealed through 18S-generated rDNA phylogeny. Mycologia. 96:773–780.
- Luangsa-Ard JJ, Hywel-Jones NL, Manoch L, Samson RA. 2005. On the relationships of Paecilomyces sect. Isarioidea species. Mycol Res. 109:581–589.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biol. 61:539–542.
- Samson RA. 1974. Paecilomyces and some allied hyphomycetes. Stud Mycol. 6:1–119.
