Abstract
Moringa oleifera is a small to medium-sized, evergreen or deciduous tree that native to northern India, Pakistan and Nepal. In this study, we report the complete chloroplast genome sequence of M. oleifera for the first time (accession number of MH778650), and it is also the first reported chloroplast genome of Moringaceae. The chloroplast genome is 160,599 bp long and includes 113 genes. Its LSC, SSC and IR regions are 88,933, 19,482 and 26,092 bp long, respectively. Phylogenetic tree analysis exhibited that M. oleifera was clustered with other Moringaceae species with 100% bootstrap values.
Moringa oleifera is a small to medium-sized, evergreen or deciduous tree that native to northern India, Pakistan and Nepal and also is known as the drumstick tree (Palada Citation1996). Not only every part of the tree be used as food, medicines or for industrial purposes, but high protein, vitamin and mineral content have made it an attractive target for wide-spread planting in some developing countries (Makkar and Becker Citation1997; Horwath and Benin Citation2011), it thus has received increased agricultural and industrial attention. In addition, M. oleifera grows well at altitudes from 0 to 1800 m and in areas with rainfall between 500 and 1500 mm per year, making it suitable for both semi-arid and arid ecosystem, which covers 37.0% of the earth’s geographical area, and even larger swaths of the developing world (Olson and Fahey Citation2011). Despite these benefits and efforts to cultivate the tree, its complete chloroplast genome was not reported although the availability of its nuclear genome sequence (Tian et al. Citation2015).
In this study, we report the complete chloroplast genome of M. oleifera in Moringaceae. DNA material was isolated from mature leaves of M. oleifera plant cultivated in the plant garden of Yunnan Institute of Tropical Crops (YITC), Jinghong, China by using DNeasy Plant Mini Kit (QIAGEN, Germany). A specimen of this tree and the isolated DNA were stored in YITC. About 10 μg isolated DNA was sent to BGI, Shenzhen for library construction and genome sequencing on the Illumina Hiseq 2000 Platform. After genome sequencing, a total of 3.8 Gbp reads in fastq format were obtained and subjected to chloroplast genome assembly. The complete chloroplast genome was annotated with Dual Organelle GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and submitted to the Genbank under the accession number of MH778650. Our assembly of the M. oleifera resulted in a final sequence of 160,599 bp in length with no gap. The overall A-T content of the chloroplast genome was 61.1%. This chloroplast genome included a typical quadripartite structure with the Large Single Copy (LSC), Small Single Copy (SSC) and Inverted Repeat (IR) regions of 88,933, 19,482 and 26,092 bp long, respectively. Genome annotation showed 113 full-length genes including 79 protein-coding genes, 30 tRNA genes and four rRNA genes. The genome organization, gene content and gene relative positions were almost identical to the previously reported Moringaceae chloroplast genomes.
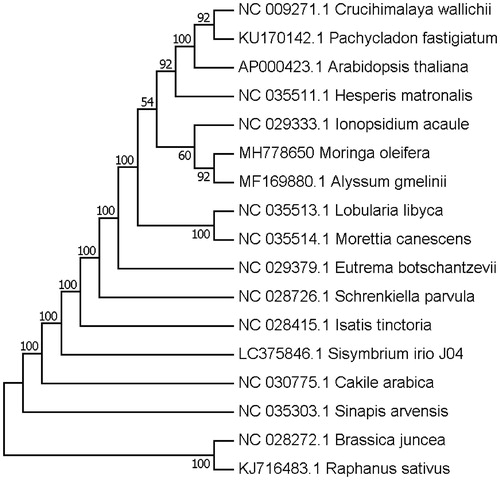
The Moringa is the only genus of Moringaceae family, and our reported M. oleifera chloroplast genome is also the first chloroplast genome in this family. To validate the phylogenetic relationships of M. oleifera, we constructed a maximum likelihood tree using 16 Brassicales taxa. The whole chloroplast genome sequences were aligned with the MAFFT (Katoh and Standley Citation2013) and the phylogenetic analysis was performed on selected taxa using RAxML (Stamatakis et al. Citation2008). The resulting tree shows that M. oleifera forms a clade with the species of Alyssum gmelinii with a 92% bootstrap value ().
Disclosure statement
The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Horwath M, Benin V. 2011. Theoretical investigation of a reported antibiotic from the “Miracle Tree” Moringa oleifera. Comput Theoret Chem. 965:196. 201
- Makkar HPS, Becker K. 1997. Nutrients and antiquality factors in different morphological parts of the Moringa oleifera tree. J Agric Sci. 128:311–322.
- Palada MC. 1996. Moringa (Moringa oleifera Lam.): a versatile tree crop with horticultural potential in the subtropical United States. Hortscience. 31:794–797.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Olson ME, Fahey JW. 2011. Moringa oleifera: a multipurpose tree for the dry tropics. Rev Mexicana Biodiversidad. 82:1071–1082.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tian Y, Zeng Y, Zhang J, Yang CG, Yan L, Wang XJ, Shi CY, Xie J, Dai TY, Peng L, et al. 2015. High quality reference genome of drumstick tree (moringa oleifera lam.), a potential perennial crop. Sci China Life Sci. 58:627–638.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255.