Abstract
Quercus variabilis plays a key role in soil remediation, erosion control, and in the economic and ecosystemic development in eastern Asia. In this study, we sequenced the complete chloroplast genome of Q. variabilis based on next-generation sequencing and used these data to assess genomic resources. The size of the Q. variabilis chloroplast genome is 161,168 bp, including a large single-copy region (90,464 bp), a small single-copy region (19,070 bp), and a pair of inverted repeats regions (25,817 bp). The overall GC content of the Q. acutissima chloroplast genome was 35.7%. The chloroplast genome of Q. variabilis encodes 112 different genes, including 78 protein-coding genes, 30 transfer RNAs (tRNA) and four ribosomal RNAs (rRNA). A phylogenetic tree constructed based on complete chloroplast genome sequences of 15 Quercus species showed that Q. variabilis was sister to Q. acutissima.
Oriental oak (Quercus variabilis) is one of the most widely distributed deciduous species in eastern Asia (Chen et al. Citation2012). It plays a key role in soil remediation, erosion control, and in the economic and ecosystemic development in China, Korea, and Japan. However, only a few genomic resources have been explored. Here, we sequenced and analyzed the chloroplast genome of Q. variabilis based on the next-generation sequencing method (Dong et al. Citation2013; Li et al. Citation2016). The objectives of this study were to establish and characterize the organization of the complete chloroplast genome of Q. variabilis and retrieve valuable genomic resources for this species.
Fresh leaves of Q. variabilis were collected from a tree in the national forest germplasm resources bank of Quercus mongolica and Quercus dentate in Hongyashan of Hebei. Total genomic DNA was extracted and purified following the method of Li et al. (Citation2013). Genomic DNA was sequenced using Illumina Hiseq 4000 PE150 (Illumina Inc., San Diego, CA). Approximately 9,910,273 paired-end reads were generated from the sequencing library. The paired-end reads were qualitatively assessed and assembled with SPAdes 3.6.1 (Bankevich et al. Citation2012). The annotation was performed with Plann (Huang and Cronk Citation2015). Finally, the whole chloroplast genome map was generated using Organellar Genome DRAW (Lohse et al. Citation2013). The sequence of Q. variabilis complete chloroplast genome was submitted to GenBank with the accession number MK105451.
The complete chloroplast genome of Q. variabilis is 161,168 bp in length. The genome's circular, quadripartite structure is composed of SSC with the length of 19,070 bp and LSC with the length of 90,464 bp, separated by a pair of IR elements (IRa and IRb) with the length of 25,817 bp. The GC content of its chloroplast DNA is 36.8%. There were 131 predicted functional genes in O. henryi chloroplast genomes, of which 113 were unique and 18 were duplicated in the IR regions. Among the 113 unique genes, there were 79 protein-coding genes, 30 tRNA genes and four rRNA genes, respectively. Most of the genes did not contain introns, 15 contained one intron, and only two contained two introns (clpP, and ycf3).
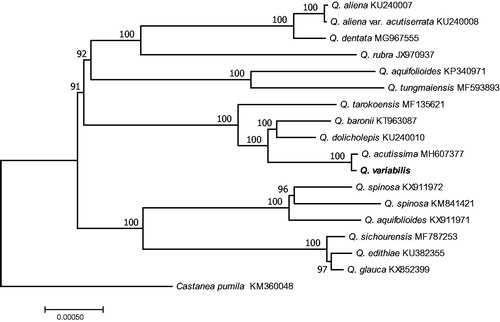
Phylogenetic analysis was reconstructed from the chloroplast genome sequences of 15 species belonging to the Quercus and Castanea pumila as an outgroup. The sequences were aligned in MAFFT v7 (Katoh and Standley Citation2013). Maximum-likelihood (ML) was used in RAxML8.0 (Stamatakis Citation2014). Relative clade support was estimated by ML bootstrap analysis of 1000 replicates of heuristic searches with the GTR + G model. ML analysis revealed that most of the nodes had 100% bootstrap values. The reconstructed phylogeny revealed that Q. variabilis was sister to Q. acutissima (). The genome information reported here could be further applied for evolution and invasion, population genetics, and molecular studies in this plant species and family.
Data availability
The chloroplast genome sequence of the Q. variabilis was submitted to Genebank of NCBI. The accession number from Genebank is MK105451.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chen D, Zhang X, Kang H, Sun X, Yin S, Du H, Yamanaka N, Gapare W, Wu HX, Liu C. 2012. Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: multiple Glacial Refugia and Mainland-Migrated Island populations. Plos One. 7:e47268.
- Dong W, Xu C, Cheng T, Lin K, Zhou S. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 5:989–997.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li B, Li Y, Cai Q, Lin F, Huang P, Zheng Y. 2016. Development of chloroplast genomic resources for Akebia quinata (Lardizabalaceae). Conserv Genetics Resour. 8:447–449.
- Li J, Wang S, Jing Y, Wang L, ZS. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48:72–78.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.