Abstract
Dumortiera hirsuta (Sw.) Nees (Dumortieraceae) is a thallose liverwort distributed primarily in tropics regions. It is the only species in family Dumortieraceae, which is a second basal family in order Marchantiales. In this study, we reported the complete chloroplast genome sequence of D. hirsuta isolated in South Korea. Its chloroplast genome was successfully assembled from raw reads sequenced by HiSeq4000. Its total length is 122,050 bp consisting of four regions: large single copy (LSC) region (81,697 bp), small single copy (SSC) region (20,061 bp), and inverted repeats (IRs; 10,146 bp per each). It contained 133 genes (89 coding genes, eight rRNAs, and 36 tRNAs); 18 genes (four rRNAs and five tRNAs) are duplicated in inverted repeat regions. The overall GC content of D. hirsuta is 28.7% and in the LSC, SSC and IR regions were 26.2%, 24.5%, and 42.8%, respectively. This chloroplast genome will be an important sequence resource for further researches of order Marchantiales.
Dumortiera hirsuta (Sw.) Nees (Dumortieraceae) is a thallose liverwort distributed in the tropical and warm temperate regions (O'Hanlon Citation1934). Based on phylogenetic analysis, it moved to Dumortieraceae from Marchantiaceae due to paraphyletic clades (Forrest et al. Citation2006; Long Citation2006), becoming the only species in that family (Forrest et al. Citation2011). Recent phylogenetic analysis presented that it is located in second basal family in order Marchantiales (Villarreal et al. Citation2016); however, it is possible to rearrange phylogenetic position of D. hirsuta because these studies used limited number of markers. Whole organelle genome sequences will provide better resolution for phylogenetic relationship. Because of previous two studies, transcriptome of D. hirsuta (Singh et al. Citation2015) and whole genome of Marchantia polymorpha subsp. ruderalis (Bowman et al. Citation2017), D. hirsuta is a suitable target for sequencing chloroplast genome as a first step to unravel phylogenetic relationship in Marchantiales.
The thallus of D. hirsuta was collected in Seogwipo, Jeju Island, in Republic of Korea and its DNA was extracted by using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Raw data generated by HiSeq4000 (Macrogen Inc, Seoul, Korea) were subject to de novo assembly done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and gap filling with SOAPGapCloser 1.12 (Zhao et al. Citation2011) to get complete chloroplast genome. Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its chloroplast genome by comparing with that of M. polymorpha subsp. ruderalis (LC192146). Voucher specimens were deposited in InfoBoss Cyber Herbarium (IN; Seoul, Republic of Korea; W., Kwon, IB-50001).
The length of D. hirsuta chloroplast genome (MH355546) is 122,050 bp and consists of large single copy (LSC; 81,697 bp) and small single copy (SSC; 20,061 bp) separated by two 10,146 bp of inverted repeats (IRs). It contains 133 genes (89 coding genes, eight rRNAs, and 36 tRNAs); nine genes (four rRNAs and five tRNAs) are duplicated in IRs. The overall GC content is 28.7% and those in the LSC, SSC, and IR regions are 26.2%, 24.5%, and 42.8%, respectively.
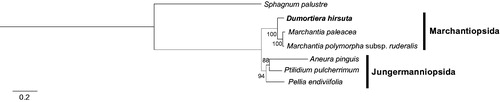
Sequence alignment of seven chloroplast genomes was conducted by MAFFT 7.388 (Katoh and Standley Citation2013). The maximum-likelihood (ML) tree was drawn using raxmlGUI 1.3 with GTR + G + I model and 1,000 bootstrap replicates (Silvestro and Michalak Citation2012). As our expectation, the phylogenetic tree presents clear distinction between two classes Marchantiopsida and Jungermanniopsida. It also shows that Dumortiera and Marchantia genera were in the same clade with bootstrap value 100 (). This chloroplast genome will be a cornerstone to understand organelle genomes in order Marchantiales.
Figure 1. Maximum-likelihood phylogenetic tree (bootstrap repeat is 1,000) of seven liverworts species based on complete chloroplast genomes: D. hirsuta (MH355546; this study), Marchantia paleacea (NC_001319), M. polymorpha subsp. ruderalis (LC192146), Pellia endiviifolia (NC_019628), Aneura pinguis (NC_035617), Ptilidium pulcherrimum (NC_015402), and Sphagnum palustre (NC_030198) as an outgroup. The numbers above branches indicate bootstrap support values.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 171:287–304.
- Forrest LL, Allen NS, Gudiño JA, Korpelainen H, Long DG. 2011. Molecular and morphological evidence for distinct species in Dumortiera (Dumortieraceae). Bryologist. 114:102–115.
- Forrest LL, Davis EC, Long DG, Crandall-Stotler BJ, Clark A, Hollingsworth ML. 2006. Unraveling the evolutionary history of the liverworts (Marchantiophyta): multiple taxa, genomes and analyses. Bryologist. 303:334.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Long D. 2006. New higher taxa of complex thalloid liverworts (Marchantiophyta–Marchantiopsida). Edinburgh J Bot. 63:257–262.
- O'Hanlon ME. 1934. Comparative morphology of Dumortiera hirsuta. Botan Gazette. 96:154–164.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organ Diversity Evol. 12:335–337.
- Singh H, Rai KM, Upadhyay SK, Pant P, Verma PC, Singh AP, Singh PK. 2015. Transcriptome sequencing of a thalloid bryophyte; Dumortiera hirsuta (Sw) Nees: assembly, annotation, and marker discovery. Sci Rep. 5:15350.
- Villarreal AJC, Crandall-Stotler BJ, Hart ML, Long DG, Forrest LL. 2016. Divergence times and the evolution of morphological complexity in an early land plant lineage (Marchantiopsida) with a slow molecular rate. New Phytol. 209:1734–1746.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
