Abstract
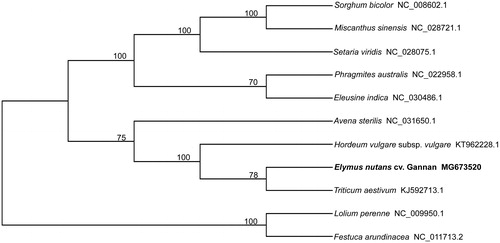
Elymus nutans is an alpine plant, which plays an important role in maintaining ecological system and the development of animal husbandry in the Tibetan region of China. However, the population of wild E. nutans notably decreased in recent years with overgrazing and ecological environment deterioration, and the germplasm resources have been threatened. This study reported a complete circular chloroplast genome of E. nutans, which was 134,147 bp in length, containing a pair of inverted repeats (IRs) of 10,541 bp separated by one small single copy (SSC) region of 14,036 bp and one large single copy (LSC) region of 99,029 bp. A total of 123 genes, including 87 protein-coding genes, 32 transfer RNA genes and four ribosomal RNA genes were predicted from the chloroplast genomes. The overall GC content of the chloroplast genome was 38.34% and the corresponding values of the IR, SSC and LSC were 48.7%, 32.4% and 37.0%, respectively. The results of phylogenetic analysis showed that wild E. nutans was clustered closely with Triticum aestivum.
The Elymus nutans belongs to Elymus of Gramineae, which mainly distributes in northwest of China with an average elevation 3000–4000 m, even exists in Himalayan area with altitude over 5200 m (Qiao et al. Citation2006). It has strong stress resistance and adaptation to cold and dry environment in Qinghai-Tibetan Plateau (QTP), which is one of the indispensable forages for yaks (Zhang et al. Citation2017). However, the rapidly increasing population of grazing yak and the destruction of ecological system caused deterioration of natural grassland, so wild E. nutans severely shrunk on the QTP. Chloroplast genomes has been widely applied in understanding the plant genetic diversity, phytogeography and conservation genetics (Burke et al. Citation2012; Zhang et al. Citation2016). In this study, we assembled and analyzed the complete chloroplast genome of Elymus nutans on Illumina Hiseq 2000 Platform (Illumina, San Diego, CA). The annotated sequence was deposited in GenBank under accession number MG673520.
Fresh leaves of wild E. nutans were collected in Maqu Tibetan Autonomous County, Gansu province, China (33°59′57′′N, 102°04′13′′E). The voucher specimens of E. nutans were deposited in the National Library of Forage Germplasm Resources in Mid-term Preservation. Genomic DNA extraction and Illumina Hiseq 2500-PE125 sequencing were conducted by Novogene Corporation (Beijing, China). Genomic alignment between the sample genome and reference genome were performed using the MUMmer (Kurtz et al. Citation2004) and LASTZ (Chiaromonte et al. Citation2002; Harris Citation2007) tools, and sample genome was annotated using the chloroplast genome of Triticum aestivum (GenBank Accession No. KJ592713.1) as a reference. Phylogenetic analysis of 11 species based on the neighbour-joining method was conducted on MEGA 6.0 (Tamura et al. Citation2013).
The complete cpDNA of wild E. nutans was 134,147 bp in length, with a pair of inverted repeat (IR) regions of 10,541 bp, a large single copy (LSC) region of 99,029 bp and a small single copy (SSC) region of 14,036 bp. It contained 123 functional genes, including 87 protein-coding genes (79 PCG species), 33 transfer RNA genes (23 tRNA species), four ribosomal RNA genes (18S and 28S rRNA) and one pseudogene (ndhF). Most gene species occur as a single copy, however, 11 PCG genes (ndhA, ndhB, ndhB1, ndhH, rps7, rps15, rpl2, rpl23, ycf2, ycf3, ycf68), seven tRNA genes (trnC-GCA, trnH-GUG, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, trnV-GAC) and two rRNA genes (18S and 28S rRNA) were duplicated. The nucleotide composition was asymmetric (30.94% A, 19.23% G, 19.11% C, 30.72% T) with an overall GC content of 38.34% and the corresponding values of the IR, SSC and LSC were 48.7%, 32.4% and 37.0%, respectively.
The phylogenetic relationships were determined with 11 cpDNA sequences of Gramineae by using the neighbor-joining method. The results showed that 11 species were clustered into two orders, and wild E. nutans was closer to Triticum aestivum (). In conclusion, the complete chloroplast genome and genomic data of wild E. nutans provide the important genetic basis for phylogenetic analysis and germplasm resources protection of wild E. nutans.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Burke SV, Grennan CP, Duvall MR. 2012. Plastome sequences of two New World bamboos – Arundinaria gigantea and Cryptochloa strictiflora (Poaceae) – extend phylogenomic understanding of Bambusoideae. Am J Bot. 99:1951–1961.
- Chiaromonte F, Yap V, Miller W. 2002. Scoring pairwise genomic sequence alignments. Pac Symp Biocomput. 7:115–126.
- Harris R. 2007. Improved Pairwise Alignment of Genomic DNA. [Ph.D. Thesis]. The Pennsylvania State University.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
- Qiao A, Han J, Gong A, Wei L, Wang Y, Qin G, Guo S, Wu J, Zhao D. 2006. Effect of nitrogen fertilizer application on Elymus nutans seed quality and yield in Qinghai-Tibet Plateau. Acta Agrestia Sinica. 14:48–47.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang D, Li K, Gao J, Liu Y, Gao L. 2016. The complete plastid genome sequence of the wild rice Zizania latifolia and comparative chloroplast genomics of the rice Tribe Oryzeae Poaceae. Front Ecol E. 4:88.
- Zhang R, Wang J, Han K, Ren T, Zeng S, Biffin E, Liu Z. 2017. Complete chloroplast genome sequence of Pedicularis cheilanthifolia, an alpine plant in China. Conserv Genet Resour. 9:1–3.