Abstract
Parasitic plants often show changes in chloroplast genome size and gene content. Here we obtained the complete chloroplast genome of Tolypanthus maclurei, a hemiparasitic shrub of Loranthaceae, using genome skimming method. It is 123,581 bp in length and contains a 72,910 bp large single copy region, a 9,731 bp small single copy region and two 20,650 bp inverted repeats. 119 genes, consisting of 77 protein-coding genes, 34 tRNA genes and 8 rRNA genes, were predicted. All the ndh genes except ndhB are lost and five protein-coding genes are pseudogenized. Phylogenetic analysis shows that Tolypanthus is sister to Macrosolen within Loranthaceae.
Parasitic plants often show more or less changes in chloroplast genome size, gene content and structure, providing insights into the evolution of plastid genomes in plants. Loranthaceae, consisting of 75 genera and about 1000 species, is a family of parasitic plants (Nickrent Citation1997). Most species of this family are aerial hemiparasites, and three other monotypic genera are entirely root parasites (Vidal-Russell and Nickrent Citation2007). So far, the chloroplast genome sequences of five species from four genera (Taxillus, Scurrula, Helixanthera and Macrosolen) of this family have been reported (Li et al. Citation2017; Shin and Lee Citation2018; Yuan et al. Citation2018). Tolypanthus, a genus of Loranthaceae, comprises only five species of hemiparasites distributed in tropical Asia (Qiu and Gilbert Citation2003). In this study, we obtained the chloroplast genome sequence of Tolypanthus maclurei (Merr.) Danser, a relatively common shrub occurring in the forests of South China, by assembling short reads produced by genome skimming.
Fresh leaves of an individual of T. maclurei were sampled in Shimentai National Nature Reserve, Yingde, Guangdong, China and the specimen was deposited at Sun Yat-sen University Herbarium (SYS). Total DNA was isolated using a modified CTAB method (Doyle and Doyle Citation1987). A shotgun genomic library with 350 bp insert size was constructed and sequenced on an Illumina HiSeq X10 platform. About 6.2 Gb paired-end, 150 bp short reads were generated and then de novo assembled into a circularized chloroplast genome with NOVOPlasty (Dierckxsens et al. Citation2017). Gene annotation was performed with manual correction in DOGMA (Wyman et al. Citation2004), setting the chloroplast genome annotation of Taxillus sutchuenensis (NC_036307) as a reference. The chloroplast genomes of 11 other species from six families of the order Santalales and Vitis rotundifolia (which was used as an outgroup) were downloaded from GenBank. MAFFT v7.311 (Katoh and Standley Citation2013) was then used for creating a multiple sequence alignment containing 49 chloroplast genes shared between T. maclurei and 11 other species of Santalales. A phylogenetic tree based on maximum-likelihood (ML) algorithm was constructed, employing RAxML v.8.0 with 1000 bootstrap replicates (Stamatakis Citation2014), to infer the phylogenetic position of T. maclurei.
The complete chloroplast genome of T. maclurei is 123,581 bp in length (GenBank accession number MH922027), which is slightly smaller relative to non-parasite flowering plants. It contains a 72,910 bp large single copy region, a 9371 bp small single copy region and two 20,650 bp inverted repeats. The overall A/T content of this chloroplast genome sequence is 63.2%. The chloroplast genome possesses 119 genes, including 77 protein-coding genes, 34 tRNA genes and eight rRNA genes. Of the 77 protein-coding genes, five (rps12, ndhB, rpl2, psbL and ycf1) are pseudogenes. All the ndh genes except ndhB, together with a tRNA gene (trnK-UUU), are found to be completely lost.
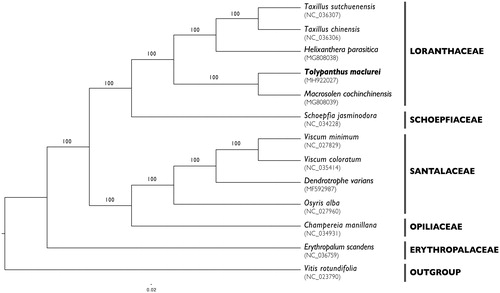
As shown in the maximum-likelihood (ML) tree (), Tolypanthus is sister to Macrosolen, another genus in Loranthaceae. The complete chloroplast genome of T. maclurei will be a useful resource for studies on phylogeny and chloroplast genome evolution in Loranthaceae.
Acknowledgements
The authors thank Yuanqiu Li at Shimentai National Nature Reserve, Yingde, Guangdong, China, for his help during our sampling.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li Y, Zhou JG, Chen XL, Cui YX, Xu ZC, Li YH, Song JY, Duan BZ, Yao H. 2017. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci Rep. 7:12834
- Nickrent DL. 1997. The Parasitic Plant Connection. Available at: < http://parasiticplants.siu.edu/> (last accessed September 14, 2018).
- Qiu HX, Gilbert MG. 2003. Loranthaceae. Flora of China, Vol. 5. Beijing: Science Press, pp. 220–239.
- Shin HW, Lee NS. 2018. Understanding plastome evolution in Hemiparasitic Santalales: Complete chloroplast genomes of three species, Dendrotrophe varians, Helixanthera parasitica, and Macrosolen cochinchinensis. PLoS One. 13:e0200293
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Vidal-Russell Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Vidal-Russell R, Nickrent DL. 2007. The biogeographic history of Loranthaceae. Darwiniana. 45:34–54.
- Yuan LX, Wang JH, Chen CR, Zhao KK, Zhu ZX, Wang HF. 2018. Complete chloroplast genome sequence of Scurrula notothixoides (Loranthaceae): a hemiparasitic shrub in South China. Mitochondrial DNA B. 3:580–581.