Abstract
Anemonefish are widely distributed in tropical areas with phenotypic color variation often observed in the same species. Complete mitochondrial genomes (mitogenomes) of 10 anemonefishes belonging to Amphiprion and Premnas were determined to support taxonomic status. Average mitogenome sequence was 16,838 ± 19.69 bp, containing 37 genes with identical gene order to most teleost mitogenomes. The percula complex comprised A. percula and A. ocellaris and was phylogenetically clustered with P. biaculeatus. Color morphs of A. ocellaris and P. biaculeatus were identified, suggesting large phenotypic variation at species level. Results will facilitate further genetic studies of mitochondrial variation and species diversity in anemonefish.
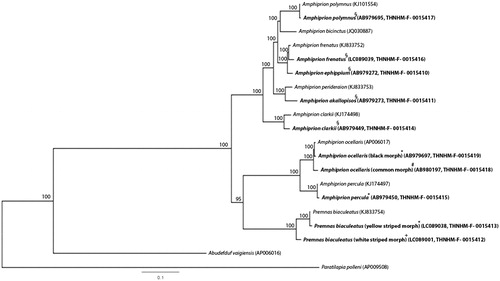
Anemonefish (Pomacentridae) are found along coral reefs. They have a mutualistic symbiotic relationship with tropical sea anemones (Elliott et al. Citation1999) and are indicators to understand the geographical distribution of high ecological speciation in the marine environment (Timm et al. Citation2008). Most are distinguished by their color patterns; however, several species show variable morphs (Allen Citation1975). Recent molecular analyses reported color patterns as not always sufficient indicators of species diversity (Li et al. Citation2015; Thongtam na Ayudhaya et al. Citation2017). Genetic diversity studies of more molecular markers will provide important information for systematic conservation anemonefish management. Here, we determined complete mitochondrial genomes (mitogenomes) of 10 anemonefishes in Amphiprion ocellaris (common morph) collected from Rayong (12.6814° N, 101.2816° E), A. ocellaris (black morph), A. percula and Premnas biaculeatus (yellow striped morph) from Chonburi, Thailand (13.3611° N, 100.9847° E), P. biaculeatus (white striped morph) from Phetchaburi, Thailand (12.9649° N, 99.6426° E), and A. clarkii, A. polymnus, A. akallopisos, A. ephippium and A. frenatus from Krabi, Thailand (8.0863° N, 98.9063° E). All specimens were stored at Thailand Natural History Museum (THNHM). Whole genomic DNA was extracted following standard salting-out protocol (Supikamolseni et al. Citation2015). Polymerase chain reaction (PCR) amplifications were performed using universal mitogenome primers (Mauro et al. Citation2004) and specific primers were developed to amplify remaining parts of the genome. All PCR products were sequenced by DNA sequencing service, First Base Laboratories Sdn Bhd (Selangor, Malaysia), with annotation performed following Srikulnath et al. (Citation2012) and Prakhongcheep et al. (Citation2018). Mitogenome sequence size of 10 anemonefishes (16,762–16,980 bp) was similar to previous reports (Mabuchi et al. Citation2007; Li et al. Citation2015; Hu et al. Citation2016; Tao et al. Citation2016). All mitogenomes comprised 37 genes and a control region (CR). Gene organization patterns were identical to teleosts (Miya et al. Citation2013). GAT deletion was found in ND5 of A. percula and A. ocellaris differed from previous reports with CCT deletion (Li et al. Citation2015), suggesting the possibility of intra-specific variation. Nucleotide diversity among anemonefish mitogenomes was 8.36 ± 0.27%, with sequence divergence between Amphiprion and Premnas at 10.98%. Two morphs of A. ocellaris and P. biaculeatus gave nucleotide divergence at 1.30% and 2.48%, respectively, similar to analysis of a single mitochondrial gene (COI, Cytb, or 16S rRNA) at species level (Thongtam na Ayudhaya et al. Citation2017), suggesting intra-specific sequence diversity. At least one tandem repeat was identified in the CR of most anemonefishes, except for A. ocellaris. The first tandem repeat was identified in Amphiprion spp., while the second in two different morphs of P. biaculeatus. A phylogenetic tree of 18 anemonefishes was constructed based on concatenated 12 protein-coding genes without ND6, with Abudefduf vaigiensis and Paratilapia polleni as the outgroup using Bayesian inference, MrBayes version 3.2.6 (Huelsenbeck and Ronquist Citation2001). All GenBank sequences were grouped within the same species, supporting Amphiprioninae as a monophyletic group (Litsios et al. Citation2014). The percula complex containing A. percula and two morphs of A. ocellaris was clustered with two morphs of P. biaculeatus (). Both morphs of A. ocellaris and P. biaculeatus were identified, suggesting large phenotypic variation at species level. Availability of complete mitogenomes will increase understanding of evolutionary processes and diversification of anemonefish and facilitate comprehensive analyses.
Figure 1 Phylogenetic relationships among concatenated mitochondrial twelve protein-coding genes, without ND6 sequences of 20 mitochondrial genomes, including Abudefduf vaigiensis and Paratilapia polleni as the outgroup using Bayesian inference analysis. The complete mitochondrial genome sequence was downloaded from GenBank. Accession and voucher numbers are indicated in parentheses after the scientific name of each species. Bold letter indicates specimens collected in this study and stored at Thailand Natural History Museum (THNHM). An asterisk (*) indicates specimens collected from Chonburi, Thailand (13.3611° N, 100.9847° E). A plus (+) indicates specimens collected from Phetchaburi, Thailand (12.9649° N, 99.6426° E). A hash (#) indicates specimens collected from Rayong, Thailand (12.6814° N, 101.2816° E). A section sign (§) indicates specimens collected from Krabi, Thailand (8.0863° N, 98.9063° E). Support values at each node are Bayesian posterior probabilities. Branch-lengths represent the number of nucleotide substitutions per site.
Acknowledgements
The authors would like to thank Sahabhop Dokkaew (Kasetsart University) for advice on sample preparation.
Disclosure statement
The authors report no conflict of interest and are entirely responsible for the content and writing of this article. Animal care and all experimental procedures were approved by the Animal Experiment Committee, Kasetsart University, Thailand (approval no. ACKU01257) and conducted in accordance with the Regulations on Animal Experiments at Kasetsart University.
Additional information
Funding
References
- Allen GR. 1975. The anemonefishes: their classification and biology. 2nd ed. New Jersey: Tropical Fish Hobbyist Publications.
- Elliott JK, Lougheed SC, Bateman B, McPhee LK, Boag PT. 1999. Molecular phylogenetic evidence for the evolution of specialization in Anemonefishes. Proc Biol Sci. 266:677–685.
- Hu X, Li J, Liu M. 2016. Complete mitochondrial genome of the pink Clownfish Amphiprion perideraion (Pisces: Perciformes, Pomacentridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1135–1136.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Li J, Chen X, Kang B, Liu M. 2015. Mitochondrial DNA genomes organization and phylogenetic relationships analysis of eight anemonefishes (Pomacentridae: Amphiprioninae). PLoS One. 10:e0123894.
- Litsios G, Pearman PB, Lanterbecq D, Tolou N, Salamin N. 2014. The radiation of the clownfishes has two geographical replicates. J Biogeogr. 41:2140–2149.
- Mabuchi K, Miya M, Azuma Y, Nishida M. 2007. Independent evolution of the specialized pharyngeal jaw apparatus in cichlid and labrid fishes. BMC Evol Biol. 7:10.
- Mauro DS, David JJG, Oommen VO, Wilkinson M, Zardoya R. 2004. Phylogeny of caecilian amphibians (Gymnophiona) based on complete mitochondrial genomes and nuclear RAG1. Mol Phylogenet Evol. 33:413–427.
- Miya M, Friedman M, Satoh TP, Takeshima H, Sado T, Iwasaki W, Yamanoue Y, Nakatani M, Mabuchi K, Inoue JG, et al. 2013. Evolutionary origin of the Scombridae (Tunas and Mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families. PloS One. 8:e73535.
- Prakhongcheep O, Muangmai N, Peyachoknagul S, Srikulnath K. 2018. Complete mitochondrial genome of mouthbrooding fighting fish (Betta pi) compared with bubble nesting fighting fish (B. splendens). Mitochondrial DNA B Resour. 3:6–8.
- Srikulnath K, Thongpan A, Suputtitada S, Apisitwanich S. 2012. New haplotype of the complete mitochondrial genome of Crocodylus siamensis and its species-specific DNA markers: distinguishing C. siamensis from C. porosus in Thailand. Mol Biol Rep. 39:4709–4717.
- Supikamolseni A, Ngaoburanawit N, Sumontha M, Chanhome L, Suntrarachun S, Peyachoknagul S, Srikulnath K. 2015. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet Mol Res. 14:13981–13997.
- Tao Y, Li J-l, Liu M, Hu X-Y. 2016. Complete mitochondrial genome of the orange clownfish Amphiprion percula (Pisces: Perciformes, Pomacentridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:324–325.
- Timm J, Figiel M, Kochzius M. 2008. Contrasting patterns in species boundaries and evolution of Anemonefishes (Amphiprioninae, Pomacentridae) in the centre of marine biodiversity. Mol Phylogenet Evol. 49:268–276.
- Thongtam na Ayudhaya P, Muangmai N, Banjongsat N, Singchat W, Janekitkarn S, Peyachoknagul S, Srikulnath K. 2017. Unveiling cryptic diversity of the anemonefish genera Amphiprion and Premnas (Perciformes: Pomacentridae) in Thailand with mitochondrial DNA barcodes. Agri Nat Resour. 51:198–205.
