Abstract
We reported the complete chloroplast genome for Zelkova serrata, the first in the distinctive genus Zelkova to be sequenced in the family Ulmaceae. The plastome is totally 158,495 bp in length, containing a pair of inverted repeats (IRs), a large single-copy region (LSR) and a small single-copy region (SSR), which are 26,456 bp, 86,921 bp and 18,684 bp, respectively. A total of 103 unique genes were identified in Z. serrata chloroplast genome, of which 88 are single-copy genes, while15 are duplicated in IR regions. Further, maximum-likelihood phylogenetic analyses were conducted using eight complete plastomes of the Ulmaceae, which support close relationships between Z. serrata, Ulmus gaussenii, and U. chenmoui.
Zelkova serrata, belonging to the genus Zelkova Spach. in the family Ulmaceae, is one of the typical deciduous broad-leaved tree species widely distributing in northern hemisphere subtropical regions (mainly in the east of China, Korea and Japan) (Kozlowski and Gratzfeld Citation2013). The species has a long history of use as a landscape tree (Fukatsu et al. Citation2012). Zelkova is a small genus containing only six species, Z. schneideriana, Z. serrata, Z. sinica (Wang et al. Citation2001), Z. carpinifolia (Czerepanov Citation1957), Z. sicula (Pasquale et al. Citation1992) and Z. abelicea (Sarlis Citation1987), and all of them are prominent Tertiary relict trees (Kozlowski et al. Citation2012). However, there have been few studies on Zelkova genetic diversity and its chloroplast genome. Here, we report the first complete chloroplast genome sequence of Zelkova.
We collected fresh leaves of Z. serrata from Anlong county of Guizhou province (25°16′52.2899N, 105°32′4.78000E) for genomic DNA extraction using the modified CTAB method (Doyle and Dickson Citation1987). The voucher specimen was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (Accession no. XTBG-BRG-SY34042). The whole chloroplast genome was sequenced following Yang et al. (Citation2014), and their nine universal primer pairs were used to perform long-range PCR for next-generation sequencing (Yang et al. Citation2014). The contigs were aligned using the publicly available cp genome of Ulmus changii (GenBank accession number KY419973) and annotated in Geneious 8.1.3.
The circular chloroplast genome is totally 158,495 bp in length, containing a pair of inverted repeats (IRs), a large single-copy region (LSR) and a small single-copy region (SSR), which are 26,456 bp, 86,921 bp and 18,684 bp, respectively. The overall G + C content was 35.6% (LSC, 33.1%; SSC, 28.5%; IR, 42.3%). A total of 103 unique genes were identified in Z. serrata chloroplast genome, of which 88 are single-copy genes, while 15 are duplicated in IR regions. Among these genes, there are 74 protein-coding genes, 25 transfer RNA (tRNA) genes, and four ribosome (rRNA) genes. In addition, seven genes (atpF, ndhA, ndhB, rpoC1, rps2, rpl2 and ycf1) contain one intron, and three genes (ycf3, rps12 and clpP) contain two introns.
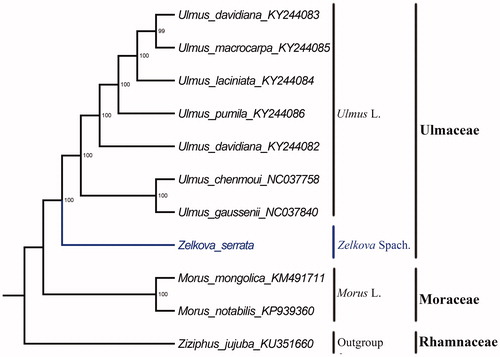
Furthermore, based on 10 published chloroplast genome sequences, we reconstructed a phylogenetic tree () to confirm the evolutionary relationship between Z. serrata and other species with published plastomes in Ulmaceae, with Ziziphus jujuba as outgroup. Maximum-likelihood (ML) (Tamura et al. Citation2011) phylogenetic analyses were performed base on GTR model in the RAxML version 8 program with 1000 bootstrap replicates (Darriba et al. Citation2012; Stamatakis Citation2014). The ML phylogenetic tree with 99% to 100% bootstrap values at each node highly supported the close relationships between Z. serrata, Ulmus gaussenii, and U. chenmoui.
Data availability
The plastome data of the Z. serrata will be submitted to Genebank of NCBI through the revision process. The accession numbers from Genebank must be supplied before the final acceptance of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Czerepanov SK. 1957. Revision specierum generum Zelkova Spach et Hemiptelea Planchon. Bot Mat Gerbariya Bot Inst V.I. Komarova Akad. Nauk SSSR. 18:58–72.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36:715–722.
- Fukatsu E, Watanabe A, Nakada R, Isoda K, Hirao T, Ubukata M, Koyama Y, Kodani J, Saito M, Miyamoto N, et al. 2012. Phylogeographical structure in Zelkova serrata in Japan and phylogeny in the genus Zelkova using the polymorphisms of chloroplast DNA. Conserv Gen. 13:1109–1118.
- Kozlowski G, Gibbs D, Fan H, Frey D, Gratzfeld J. 2012. Conservation of threatened relict trees through living ex situ collections: lessons from the global survey of the genus Zelkova (Ulmaceae). Biodiversity Conserv. 21:671–685.
- Kozlowski G, Gratzfeld J. 2013. Zelkova – an ancient tree. Global status and conservation action. Switzerland: Natural History Museum Fribourg.
- Pasquale GD, Garfi G, Quézel P. 1992. Sur la présence d'un Zelkova nouveau en Sicile sud-orientale (Ulmaceae). (On the occurrence of a new species of Zelkova in south eastern Sicily.). Biocosme Mésogéen. 8:401–409.
- Sarlis G. 1987. Zelkova abelicea (Lam.) Boiss., an endemic species of Crete (Greece). Webbia. 41:247–255.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wang YF, Ferguson DK, Zetter R, Denk T, Garfi G. 2001. Leaf architecture and epidermal characters in Zelkova, Ulmaceae. Bot J Linnean Soc. 136:255–265.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.