Abstract
Hemiptelea davidii (Hance) Planch is a potential valuable forest tree in China. Here, the complete chloroplast genome of H. davidii was reported using high-throughput sequencing technique. The chloroplast genome is 160,195 bp in length, contains a pair of inverted repeat regions (IR, 26,599 bp), which were separated by a large single copy region (LSC, 88,231 bp) and a small single copy region (SSC, 18,766 bp). The GC content of the whole chloroplast genome is 35.28%. The chloroplast genome comprises 111 unique genes, including 77 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analyses of 17 chloroplast genomes indicated that H. davidii is close with Zelkova and Ulmus species in Ulmaceae but little far from the species in Cannabaceae, Moraceae and Urticaceae.
Hemiptelea davidii (Hance) Planch is the only species of the genus Hemiptelea of Ulmaceae, which is mainly distributed in China. It shows strong resistance to drought, cold, and saline/alkali condition, and is then recognized as a valuable forest tree resource for arid sandy environments (Bai et al. Citation2008). Also, H. davidii could be used as a potential medicinal plant since of the rich flavonoids substances inside (Ma et al. Citation2013). Unfortunately, researches regarding its genetic background are extremely scarce. In this study, we first reported the complete chloroplast genomes for H. davidii based on high-throughput sequencing technique, so as to well understand their characteristics and potential application. The annotated chloroplast genome of H. davidii has been deposited into GenBank under accession number MK070168.
The plant material was collected in Yantai of Shandong Province, China. The voucher specimens were deposited in Shandong Provincial Center of Forest Tree Germplasm Resources. The total genomic DNA of H. davidii was extracted using the Plant Genomic DNA Kit, and sequenced by Illumina Hiseq2500 System. The sequences were assembled by SOAPdenovo software (Luo et al. Citation2012), and further screened based on the known chloroplast reference genome. The chloroplast genome sequences were annotated with CpGAVAS (Liu et al. Citation2012) and DOGMA (Wyman et al. Citation2004).
The complete chloroplast genome of H. davidii is 160,195 bp in length, forming a typical circular quadripartite structure with a pair of inverted repeat regions (IR, 26,599 bp) separated by a large single copy region (LSC, 88,231 bp) and a small single copy region (SSC, 18,766 bp). The GC contents of the whole chloroplast genome are 35.28%. The chloroplast genome comprises of 111 unique genes, including 77 protein-coding genes, 30 tRNA genes, and four rRNA genes. Fourteen genes have a single intron, three genes (ycf1, ycf3, and clpP) have two introns, and the others have no introns. Most of the genes occurred in a single copy but 19 genes were duplicated in the IR regions, including eight protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12, and ycf1), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5).
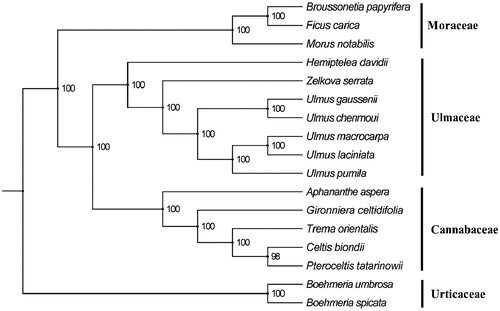
The whole of chloroplast genome sequence were aligned by MAFFT (Katoh and Standley Citation2013). Phylogenetic analyses were performed using maximum likelihood (ML) with RAxML based on GTRGAMMA model with 1000 bootstrap replicates (Silvestro and Michalak Citation2012). As shown in ML phylogenetic tree (), the species in Ulmaceae formed a monophyletic clade with a high bootstrap value (100%), and distinguished with the species in Cannabaceae that were recognized as Ulmaceae plants traditionally. The complete chloroplast genome reported here will provide a useful resource for further exploring the phylogeny, population genetic diversity and conservation genetics of H. davidii.
Figure 1. The phylogenetic tree based on 17 complete chloroplast genomes. Accession number: Broussonetia papyrifera (KX828844), Ficus carica (KY635880), Morus notabilis (KP939360), Hemiptelea davidii (MK070168), Zelkova serrata (AP017905), Ulmus gaussenii (MG599732), Ulmus chenmoui (MG581403), Ulmus macrocarpa (KY244085), Ulmus laciniate (KY244084), Ulmus pumila (KY244086), Aphananthe aspera (AP017911), Gironniera celtidifolia (KY931662), Trema orientalis (KY931668), Celtis biondii (MH118119), Pteroceltis tatarinowii (MH118125), Boehmeria umbrosa (MF990291), and Boehmeria spicata (MF990290). The number on each node indicates the bootstrap value.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bai YP, Han DY, Dong YH, Zhao YJ, Li JD. 2008. Structural characteristics of Hemiptelea davidii community on Kerqin sandy land. Chinese J Appl Ecol. 19:257–260.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, 2012. Soapdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18.
- Ma FF, Shi LC, Bao HY. 2013. Study on quality evaluation of different parts from Hemiptelea davidii and its antioxidant activity. Ginseng Res. 25:19–23.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
