Abstract
In this study, we report the first complete mitochondrial genome of three Mesabolivar specimens found in the interior of N4E_0023 cave from Serra Norte (Carajás), Parauapebas (Brazil). The three mitogenomes contain 14,941, 14,845 and 14,727 bp, and GC content of 29.41%, 31.68% and 29.34%, respectively. All three mitogenomes include 13 protein-coding genes, 17 transfer RNA (tRNA) genes, five putative transfer RNA (tRNA) genes and two ribosomal RNA (16S and 12S rRNA). We also performed a phylogenetic analysis with the concatenated coding genes from the complete mitochondrial genomes and showed that the analyzed Mesabolibar specimens clustered together in a clade, sister to the group with two Pholcus species, the other Pholcidae species with available mitogenome.
The genus Mesabolivar was first described by González-Sponga (Citation1998) and includes 94 species (Huber Citation2018), is widespread in the Neotropical regions (Machado et al. Citation2007) and one of the most species-rich pholcid genus. Identification of Mesabolivar in the field is difficult, considering their diversity in morphological and behaviour traits (Huber Citation2018). The three troglophile specimens of Mesabolivar were collected from the interior of N4E_0023 cave located in Serra Norte, mining province of Carajás, Parauapebas, Brazil. Here we report the first complete mitogenomes for Mesabolivar species, which could facilitate species determination and genetic studies of the taxon.
Genomic DNA of the samples was isolated using the DNeasy Blood & Tissue Kit (Qiagen), following the manufacturer’s protocol for insects, and are stored at ITV (Instituto Tecnológico Vale) under the codes ITV1036I1, ITV1036I2 and ITV1036I3. Paired-end libraries were constructed from approximately 50 ng of DNA using the QXT SureSelect (Agilent Technologies) kit according to the manufacturer’s instructions. The sequencing run was performed in the Illumina NextSeq 500 platform (300 cycles). The mitogenome assembly was performed with NovoPlasty 2.6.7 (Dierckxsens et al. Citation2016) and the annotations with Mitos (Bernt et al. Citation2013).
The complete mitogenomes of the three Mesabolivar species (GenBank accessions MH643812, MH643813, and MH643814) had a length of 14,941, 14,845 and 14,727 bp, and GC content of 29.41%, 31.68% and 29.34%, respectively. All three genomes included 13 protein-coding genes, 17 tRNA genes, five putative tRNA genes, and the two rRNA (16S and 12S) genes. In the three genomes, all genes except NAD1, NAD4, NAD4L, NAD5 and nine tRNA were encoded on the L-strand. The gene COX1 of MH643812 and MH643814 were identical, indicating that they are the same species (Mesabolivar sp.1), while the considerably lower similarity with MH643813 (84.1%) denotes another species (Mesabolivar sp.2). Mesabolivar sp.1 had five ORFs starting with ATG, four starting with ATT, NAD1 with ATC, NAD6, and COX3 with ATA, and COX1 with TTG. Mesabolivar sp.2 had four ORFs starting with ATA, four with TTG, three with ATG, NAD1 with ATC, and COX2 with GTG. Regarding the stop codon, eight genes ended with TAA, COX1 and ATP6 with TAG, ATP8 with TA, and COX2 and NAD4L with T in sp.1, while in sp.2, 11 genes ended with TAA and two genes ended with TAG. The base composition of protein-coding genes for sp.1 and sp.2 were, respectively, A in 29.4% and 28.0%, C in 13.8% and 14.5%, G in 17.0% and 18.1%, and T in 39.9% and 39.3%.
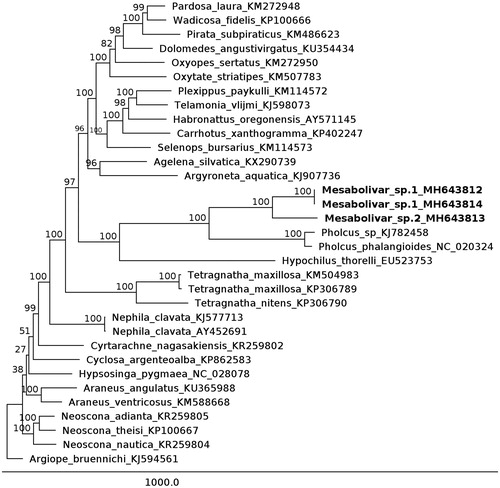
For the phylogenetic analysis, 30 additional mitogenomes of Araneomorphae species were obtained, and the coding genes were aligned using MAFFT v7.271 (Katoh et al. Citation2002). We used RaxML v8.2 (Stamatakis Citation2014), as implemented in the CIPRES portal (http://www.phylo.org), to perform a maximum likelihood analysis, with 1000 bootstrap replications, using the GTR-G model. Argiope bruennnichi (KJ594561) served as the outgroup. The phylogenetic tree showed that the Mesabolivar sp.1 specimens were grouped as sister to Mesabolivar sp.2 and related to the other mitogenomes from the Pholcidae family, as expected ().
Disclosure statement
No potential conflict of interest was reported by the authors. The results of this work were not directly used for any environmental licensing purposes. There are no patents, products in development or marketed products to declare.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:gkw955.
- González-Sponga MA. 1998. Arácnidos de Venezuela. Cuatro nuevos géneros y cuatro nuevas especies de la familia Pholcidae Koch, 1850 (Araneae). Mem Fund La Salle Cien Nat. 57:17–31.
- Huber BA. 2018. The South American spider genera Mesabolivar and Carapoia (Araneae, Pholcidae): new species and a framework for redrawing generic limits. Zootaxa. 4395:1.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Machado ÉO, Brescovit AD, Candiani DF, Huber BA. 2007. Three new species of Mesabolivar (Aranea, Pholcidae) from leaf litter in urban environments in the city of São Paulo, São Paulo, Brazil. Iheringia Série Zool. 97:168–176.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.