Abstract
Marula, Sclerocarya birrea (A. Rich.) Hochst. subsp. Caffra (Sond.) Kokwaro, is an indigenous fruit tree species of southern Africa with considerable socioeconomic importance. The species belongs to the Anacardiaceae family and Sapindales. Here we sequenced and assembled the complete plastid genome of marula. The marula plastome is 162,517 bp in length and contains a pair of inverted repeat (IR) regions of 26,790 bp each, separated by a large single-copy (LSC) region of 89,874 bp and a small single-copy (SSC) region of 19,063 bp. It encodes 78 protein-coding genes, 28 tRNA genes, and four rRNAs. We performed Maximum-likelihood (ML) phylogenetic analysis based on 80 plastid genes from 53 Sapindales species, and the result supported the recent treatment of Sapindales and the placement of marula in Anacardiaceae.
Sclerocarya birrea (A. Rich.) Hochst. subsp. Caffra (Sond.) Kokwaro, known as marula, is a traditionally important African indigenous tree in southern Africa (Emanuel et al. Citation2005), and plays an integral part in the lives, food security and spirituality of the local communities in the Bushbuckridge Lowveld, South Africa (Shackleton and Shackleton Citation2000). Marula has a wide variety of usages, including using fresh fruits for direct consumption or beverages, kernels as food, flavour, and preservatives, bark as herb medicine, and wood as firewood and carvings (Shackleton and Shackleton Citation2000). Despite its considerable socioeconomic importance, little is known regarding its genetic background.
Plastid DNA has provided important knowledge in plant ecology and genetics (Twyford and Ness Citation2017) and allows for a better understanding of these important crop species. Here we assembled and described the complete plastid genome of Sclerocarya birrea, the marula tree using the NGS technology. The annotated genome sequence has been deposited in the GenBank database (MK002721).
Fresh marula leaves were collected from an individual tree at the World Agroforestry Center (ICRAF) at Nairobi, Kenya. Genomic DNA was extracted using the CTAB method (Lodhi et al. Citation1994), and stored (R0141201631) at the China National GeneBank at Shenzhen, China. NGS libraries with 250 bp insert size was prepared and sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA), yielding 40 Gb high quality reads. After trimming with SOAPfilter v2.2 using default parameters, 240 M reads were kept. Plastid genome was assembled using NOVOPlasty v2.5.9 (Dierckxsens et al. Citation2017) with Arabidopsis thaliana plastome (NC_000932.1) as the seed sequence and annotated in Geneious 10.0.0 (https://www.geneious.com/) with the plastome of Spondias mombin (KY828469) as the reference.
The complete plastid genome of marula is a circular, double-stranded DNA molecule of 162,517 bp, with two inverted repeats (IRs) of 26,790 bp separated by a large single-copy (LSC) region of 89,874 bp and a small single-copy (SSC) region of 19,063 bp. It encodes 78 protein-coding genes (PCGs), 28 tRNA genes and four rRNAs. A total of 19 genes are duplicated in the IR regions, including eight PCGs (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1 and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC), and all four rRNA genes (rrn4.5, rrn5, rrn16 and rrn23). Seven PCGs (atpF, ndhA, ndhB, rpl2, rpoC1, rps16 and rps19) and four tRNA genes (trnA-UGC, trnI-GAU, trnL-UAA and trnV-UAC) harbor a single intron, and three genes (clpP, rps12 and ycf3) possess two introns.
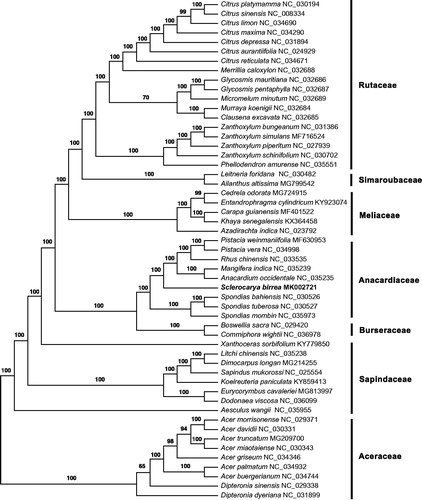
To assess the phylogenetic position of marula within the Sapindales, we constructed a concatenated data matrix based on the 80 plastid genes including 52 published plastomes of Sapindales from the NCBI database. Individual gene was aligned using the program MAFFT v7.017 (Katoh and Standley Citation2013), and concatenated into a data matrix of 67,389 sites. The Maximum-Likelihood tree was constructed under the GTRCAT model using RAxML v8.2.4 (Stamatakis Citation2014) with 100 bootstrap replicates. Maximum likelihood analysis supports the current treatment of the order Sapindales (Chen Citation2018) and the placement of S. birrea in the Anacardiaceae clade ().
Acknowledgements
The authors thank Xuezhu Liao at BGI, Shenzhen for the assistance with the data analyses.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen KK. 2018. Characterization of the complete chloroplast genome of the tertiary relict tree Phellodendron amurense, (sapindales: rutaceae) using illumina sequencing technology. Conserv Genet Resour. 10:43–46.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Emanuel PL, Shackleton CM, Baxter JS. 2005. Modelling the sustainable harvest of Sclerocarya birrea subsp. caffra fruits in the South African Lowveld. Forest Ecol Manag. 214:91–103.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lodhi MA, Ye GN, Weeden NF, Reisch BI. 1994. A simple and efficient method for DNA extraction from grape vine cultivars and Vitis species. Plant Mol Biol Report. 12:6–13.
- Shackleton CM, Shackleton SE. 2000. Direct-use values of secondary resources harvested from communal savannas in the Bushbuckridge lowveld, South Africa. J Trop For Prod. 6:28–47.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Twyford AD, Ness RW. 2017. Strategies for complete plastid genome sequencing. Mol Ecol Resour. 17:858–868.