Abstract
Bennettiodendron brevipes, which is only distributed in China, is a genus of the family Salicaceae. In this study, we assembled the complete chloroplast genome of B. brevipes using Illumina paired-end sequencing data. The complete chloroplast genome sequence of B. brevipes is 158,605 bp in size. It is composed of a large single-copy (LSC), a small single-copy (SSC), and two inverted repeat (IR) regions of 86,350, 16,501, and 22,877 bp, respectively. In addition, the overall GC-content of the whole plastome is 36.66%. The B. brevipes chloroplast genome has a total of 126 genes including 82 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Furthermore, phylogenetic analysis based on the chloroplast genome demonstrated that B. brevipes is most closely related to Idesia polycarpa.
Keywords:
Bennettiodendron brevipes, which was included in the family Flacourtiaceae, but now belongs to the family Salicaceae (Chase et al. Citation2002). This small evergreen tree is only distributed in several provinces of southern China, such as Hunan, Jiangxi, Guangdong, Guizhou, and Yunnan and it grows at elevations of 400–1800 m. Most genera of Salicaceae have genome data of nucleus or organelle (Ma et al. Citation2013; Chen et al. Citation2016; Zhang and Gao Citation2016; Yang et al. Citation2017), however, there is no sequence data of B. brevipes. In order to get a further understanding of Salicaceae phylogenetic analysis, assembling a complete plastome of B. brevipes is necessary and it will be helpful to raise our awareness of family Salicaceae and B. brevipes. Therefore, we assembled and obtained the complete chloroplast genome of B. brevipes using Illumina paired-end sequencing data.
The voucher specimen of B. brevipes was stored at Herbarium of Arnold Arboretum, Harvard University. Fresh leaves were collected from Huaiji, Guangdong province, China and the modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987) was used to extract the total genomic DNA. The whole-genome sequencing was conducted with 350 bp pair-end reads on the Illumina Hiseq platform (Illumina, San Diego, CA). As a result, we obtained high quality clean reads which were used for the cp genome assembly. The programme NOVO-Plasty (Dierckxsens et al. Citation2017) was used to assemble the cp genomes. The plastome was annotated using Plann (Huang and Cronk Citation2015). Next, we corrected the annotation with Geneious (Kearse et al. Citation2012) and we chose plastid genomes of 15 species to do alignments by MAFFT (Katoh and Standley Citation2013). With 100 bootstrap replicates and the alignments as an input file of RAxML version 8 (Stamatakis Citation2014), we obtained a neighbour-joining (ML) tree. The B. brevipes whole chloroplast genome sequence was deposited to GenBank® under accession number: MK046729.
After manual adjustment, we obtained a 158,605 bp complete cp genome of B. brevipes, whose length is near to other Salicaceae chloroplast genome levels (Kersten et al. Citation2016). The base composition of the circular cp genome was asymmetric (31.31% A, 18.66% C, 18.00% G, and 32.02% T). The plastome, which was separated by two inverted repeat (IR) regions of 55,754 bp contains a large single-copy (LSC) region of 86,350 bp and a small single-copy (SSC) region of 16,501 bp. The overall GC content of Camptotheca acuminata cpDNA is 36.66%, while the corresponding values of the LSC, SSC, and IR regions are 34.4, 30.68, and 41.88%, respectively. The cp genome had 126 genes, including 82 protein-coding genes, 36 transfer RNA genes, and eight ribosomal RNA genes.
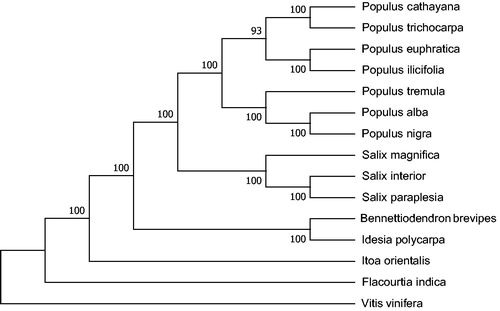
The phylogenetic analysis of 15 plastid genomes, which Vitis vinifera was used as outgroup, showed that the cp genome of B. brevipes is closely related to the species of Idesia polycarpa ().
Figure 1. The phylogenetic tree based on the 13 complete chloroplast genome sequences. Accession numbers: Populus alba (AP008956.1), Populus cathayana (KP729175.1F), Idesia polycarpa (KX229742.1), Populus ilicifolia (KX421095.1), Vitis vinifera (NC_007957.1), Populus trichocarpa (NC_009143.1), Salix interior (NC_024681.1), Populus euphratica (NC_024747.1), Populus tremula (NC_027425.1), Populus nigra (NC_037416.1), Salix magnifica (NC_037424.1), Salix paraplesia (NC_037426.1), Flacourtia indica (NC_037410.1), Itoa orientalis (NC_037411.1), and Bennettiodendron brevipes (MK046729).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Chase MW, Zmarzty S, Lledo MD, Wurdack KJ, Swensen SM, Fay MF. 2002. When in doubt, put it in Flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull. 57:141–181.
- Chen Z, Wang W, Yang W, Ma T. 2016. Characterization of the complete chloroplast genome of Populus ilicifolia. Conserv Genet Resour. 8:391–393.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kersten B, Faivre Rampant P, Mader M, Le Paslier MC, Bounon R, Berard A, Vettori C, Schroeder H, Leplé JC, Fladung M, et al. 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLoS One. 11:e0147209.
- Ma T, Wang J, Zhou G, Yue Z, Hu Q, Chen Y, Liu B, Qiu Q, Wang Z, Zhang J, et al. 2013. Genomic insights into salt adaptation in a desert poplar. Nat Commun. 4:2797.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yang W, Wang K, Zhang J, Ma J, Liu J, Ma T. 2017. The draft genome sequence of a desert tree Populus pruinosa. Gigascience. 6:1–7.
- Zhang Q, Gao L. 2016. The complete chloroplast genome sequence of desert poplar (Populus euphratica). Mitochondrial DNA A DNA Mapp Seq Anal. 27:721–723.
