Abstract
In this study, the complete chloroplast genome sequence of Podocarpus imbricatus (Podocarpaceae) was determined. Like other chloroplast genomes in conifers, the Podocarpus imbricatus genome did not have typical quadripartite structure. The genome was 133,816 bp in length, containing 118 unique genes (82 protein-coding genes, 32 tRNA genes and four rRNA genes). The overall GC content of the genome is 37.23%. A phylogenetic tree based on 56 chloroplast genes showed that Podocarpaceae was clustered with Araucariaceae. In addition, all the Podocarpaceae species analyzed in this study formed a monophyletic clade.
Podocarpus imbricatus, belongs to the Podocarpaceae family, is mainly distributed in Southern China, Myanmar, Vietnam and Indonesia. The timber of P. imbricatus is used for construction and making furniture. The 20-Hydroxyecdysone isolated from P. imbricatus bark can inhibit the proliferation of acute myeloid leukaemia cells (Thuy et al. Citation2017). However, in the past years, P. imbricatus has experienced indiscriminate felling and then was listed as threatened plant in the China Plant Red Data Book (Fu and Jin Citation1992). The Podocarpaceae is the largest family in cupressophytes, comprising about 19 genera with 180 species (Christenhusz et al. Citation2011). To date, the complete chloroplast genome from 13 species covering 12 genera in this family has been reported (Vieira et al. Citation2014; Wu and Chaw Citation2014, Citation2016; Sudianto et al. Citation2018). In order to have a deep insight into the evolution of Podocarpaceae, the complete chloroplast genome sequence of P. imbricatus was sequenced in this study.
The fresh leaves of P. imbricatus used in this study were collected from South China Botanical Garden, Chinese Academy of Sciences. The voucher specimens of P. imbricatus were deposited in Shaanxi Xueqian Normal University. Total genomic DNA was extracted using the modified CTAB method (Gawel and Jarret Citation1991) and sequenced by the Illumina HiseqTM 2500 platform (Illumina Inc., San Diego, CA). The complete chloroplast genome sequence of P. imbricatus was assembled by NOVOPlasty software (Dierckxsens et al. Citation2017). Annotations were originally performed by DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). The exact boundaries of each gene were verified by alignment with other conifers chloroplast gene.
The gene order and structure of P. imbricatus chloroplast genome are similar to other conifer species, which did not have the typical inverted repeat sequence. The complete chloroplast genome sequence of P. imbricatus is 133,816 bp in length (GenBank accession no. MH979330). In total, 118 unique genes were identified, including 82 protein-coding genes, 32 tRNA genes and four rRNA genes. Fourteen genes contain one intron and ycf3 has two introns. In addition, rps16 gene was lost and two copies of trnN-GUU gene were found in P. imbricatus chloroplast genome. The overall GC content of the chloroplast genome is 37.23% (protein-coding genes, 38.18%; tRNA genes, 53.01%; rRNA genes, 54.03%; introns, 37.84%; intergenic spacers, 33.70%). The percentage of the coding region is 55.40% and the gene density is 0.89 genes/kb.
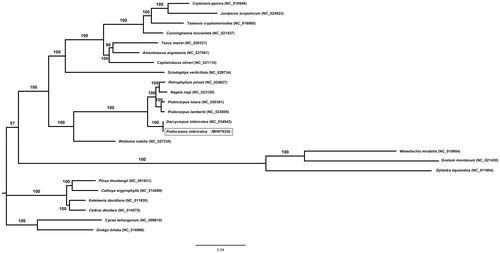
Maximum likelihood phylogenetic tree was constructed by 52 protein-coding and four rRNA genes from 24 gymnosperm taxa, using Cycas taitungensis and Ginkgo biloba as the outgroup. The dataset was implemented in RAxML v8.1.x (Stamatakis Citation2014). The ML tree strongly supported the sister relationship between P. imbricatus and a clade containing Podocarpus totara, Podocarpus lambertii, Nageia nagi and Retrophyllum piresii. In addition, Podocarpaceae was clustered with Wollemia nobills (Araucariaceae) by 100% bootstrap (). The complete chloroplast genome sequence of P. imbricatus facilitates a better understanding of population and phylogenetic studies for Podocarpaceae.
Disclosure statement
The author declares no conflicts of interest. The author alone is responsible for the content and writing the manuscript.
Additional information
Funding
References
- Christenhusz MJM, Reveal JL, Farjon A, Gardner MF, Mill RR, Chase MW. 2011. A new classification and linear sequence of extant gymnosperms. Phytotaxa. 19:55–70.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Fu LK, Jin JM. 1992. China plant red data book: rare and endangered plants I. Beijing: Science Press. p. 132–133.
- Gawel N, Jarret R. 1991. A modified CTAB DNA extraction procedure for musa and ipomoea. Plant Mol Biol Rep. 9:262–266.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sudianto E, Wu CS, Leonhard L, Martin WF, Chaw SM. 2018. Enlarged and highly repetitive plastome of Lagarostrobos and plastid phylogenomics of Podocarpaceae. bioRxiv.
- Thuy TT, Tam NT, Anh NT, Hau DV, Phong DT, Thang LQ, Adorisio S, Van Sung T, Delfino DV. 2017. 20-Hydroxyecdysone from Dacrycarpus imbricatus bark inhibits the proliferation of acute myeloid leukemia cells. Asian Pac J Trop Med. 10:157–159.
- Vieira LN, Faoro H, Rogalski M, Fraga HP, Cardoso RL, de Souza EM, de Oliveira Pedrosa F, Nodari RO, Guerra MP. 2014. The complete chloroplast genome sequence of Podocarpus lambertii: genome structure, evolutionary aspects, gene content and SSR detection. PLoS One. 9:e90618.
- Wu CS, Chaw SM. 2014. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol J. 12:344–353.
- Wu CS, Chaw SM. 2016. Large-Scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (cupressophytes). Genome Biol Evol. 8:3740–3750.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.