Abstract
The complete mitochondrial genome of a specimen of Cydnidae was shotgun sequenced and each partition was characterized. This genome, with 15,289 bp in length, has all of the 37 genes commonly found in metazoan mitochondrial genomes: two for ribosomal RNAs (rRNAs), 22 for transfer RNAs (tRNAs), and 13 for proteins. Protein-coding and ribosomal genes have a similar arrangement as in other insects. Phylogenetic relationship of this species with other family groups within the infraorder Pentatomomorpha was inferred using the maximum likelihood method based on the mitogenome. The phylogenetic analysis groups this specimen with the other Cydnidae species within the Pentatomoidea clade.
The Cydnidae are the second most speciose family within Pentatomoidea (Hemiptera: Heteroptera: Pentatomomorpha). Commonly known as Burrower Bugs, they are mostly black and brown insects which spend most of the time buried in the soil (Schwertner and Nardi Citation2015). Six subfamilies are recognized within Cydnidae and its monophyly is controversial (Lis Citation2010). In tropical caves, large populations of Cydnidae bugs are frequently found breeding and feeding on guano of frugivorous bats (Gnaspini and Trajano Citation2000). Molecular studies are still scarce, with just one representative with the mitogenome sequenced (Hua et al. Citation2008). Among with the limited taxonomic identification, and the lack of molecular information, the relationship between the Cydnidae are barely understood. Sequencing the mitogenome of Cydnidae species is essential to understand the phylogenetic relationship of the family. In this study, the mitochondrial genome of a Cydnidae was sequenced and annotated and the results were used to confirm the phylogenetic position of this specimen.
A troglophile Cydnidae was used for whole-genome shotgun sequencing and sequence assembly de novo. The specimen was collected at N4E_0023 cave located in Parauapebas, Pará, Brazil. Genomic DNA was extracted using the DNA Blood & Tissue Kit (Qiagen, Hilden, Germany) following the insect tissue protocol and is deposited at Instituto Tecnológico Vale (ITV) under the code ITV1034. DNA library was constructed with QXT SureSelect Kit (Agilent Technologies, Santa Clara, CA) and sequenced with NextSeq 500 platform (Illumina, San Diego, CA). The mitochondrial sequences were assembled with NovoPlasty software version 2.6.7 (Dierckxsens et al. Citation2017), annotated using the MITOS web server (Bernt et al. Citation2013) and deposited in GenBank under accession code MH643815.
The mitogenome of Cydnidae sp. is a circular molecule of 15,289 bp in length and has all the 37 commonly found genes between the animal mitochondrial genomes (Boore Citation1999) which consists of 13 protein-coding genes (PCG), 22 transfer RNAs (tRNAs), and 2 ribosomal RNAs (rRNAs). The base composition of the mitogenome was estimated to be A 41.3%, T 32.28%, C 15.25%, G 10.82% with a high A-T content (73.9%). With exception of NAD1, NAD4, NAD4L, NAD5, eight tRNAs, and the two rRNAs, all genes are coded in the H-strand. The start codon of most PCGs is ATN except for COX1 (TTG), ATP8 (GTG), and NAD1 (GTG), as observed in other pentatomoid mitogenomes (Yuan et al. Citation2015). For the stop codon, most PCGs ended with TAN, but truncated stop codons were also found for COX2, COX3, ATP6, and NAD5.
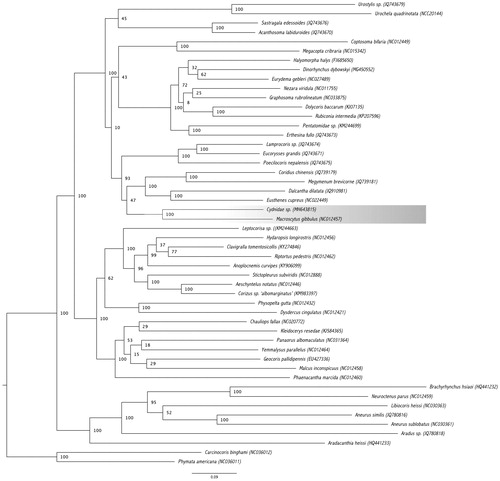
We compared our mitogenome sequence to the other 47 Pentatomomorpha mitochondrial sequences available in GenBank in a maximum likelihood phylogenetic analysis (). Two Cimicomorpha species were used as the outgroup, Phymata americana, and Carcinocoris binghami. The nucleotide sequences of 13 PCG and two rRNAs were extracted using Geneious and subsequently aligned using MAFFT version 7.271 (Katoh and Standley Citation2013). The resulting alignments were concatenated into a nucleotide matrix which was used in the phylogenetic analysis with RAxML version 8.2.10 (Stamatakis Citation2014) via Cipres portal (www.phylo.org). Our Cydnidae specimen clustered with the other available Cydnidae mitogenome, Macroscytus gibbulus within the Pentatomoidea clade.
Disclosure statement
The results of this work were not directly used for any environmental licensing purposes. There are no patents, products in development or marketed products to declare.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Gnaspini P, Trajano E. 2000. Guano communities in tropical caves. In Wilkins H, Culver DC, Humphreys WF, editors. Subterranean ecosystems. Amsterdam: Elsevier Press; p. 251–268.
- Hua J, Li M, Dong P, Cui Y, Xie Q, Bu W. 2008. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genom. 9:610.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lis JA. 2010. Coxal combs in the Cydnidae sensu lato and three other related “cydnoid” families – Parastrachiidae, Thaumastellidae, Thyreocoridae (Hemiptera: Heteroptera): functional, taxonomic, and phylogenetic significance. Zootaxa. 2476:53–64.
- Schwertner CF, Nardi C. 2015. Burrower bugs (Cydnidae). In: Panizzi AR, Grazia, J, editors. True bugs (Heteroptera) of the neotropics. Entomology in focus. Dordrecht, Netherlands: Springer; p. 639–680.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yuan M, Zhang Q, Guo Z, Wang J, Shen Y. 2015. Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications. BMC Genom. 16:460.