Abstract
Nymphaea lotus L. (Nymphaeaceae) is a type species of subgenus Lotos, which has been frequently cultivated in ornamental garden pools. Here, we presented complete chloroplast genome of N. lotus which is 159,311 bp long and has four subregions: 89,610 bp of large single copy (LSC) and 19,333 bp of small single copy (SSC) regions are separated by 25,184 bp of inverted repeat (IR) regions including 130 genes (85 coding genes, eight rRNAs, and 37 tRNAs). The overall GC contents of the chloroplast genome were 39.1% and in the LSC, SSC, and IR regions were 37.7%, 34.2%, and 43.4%, respectively. Phylogenetic tree constructed with whole chloroplast genomes present phylogenetic position of subgenus Lotos.
Nymphaea lotus L. which is a type species of subgenus Lotos lives in various parts of East Africa and Southeast Asia (Conard Citation1905). Due to its beautiful flowers, it has been utilized as ornamental aquatic plant species in garden. Nymphaea species in subgenus Lotos usually bloom from sunset to next morning in tropical area (Hirthe and Porembski Citation2003). Based on phylogenetic study with trnT–trnF sequence on chloroplast, subgenus Lotos was located at second basal clade after subgenus Nymphaea (Borsch et al. Citation2007). Based on available chloroplast genomes of genus Nymphaea, there is no available chloroplast genome in subgenus Lotos.
Nymphaeaceae family containing N. lotus is one of basal angiosperm families after the most basal family, Amborellaceae family in which Amborella trichopoda genome was sequenced to understand evolutionary history of early Angiosperms (Albert et al. Citation2013). A next target to dissect whole genome sequences to understand evolutionary history of early Angiosperms will be one of species in Nymphaeaceae: Nymphaea colorata (Chen et al. Citation2017) and Nymphaea capensis (unpublished, InfoBoss Co., Ltd., Seoul, Korea) genomes are being sequenced.
Here, we sequenced complete chloroplast genome of N. lotus (Voucher deposited in the InfoBoss Cyber Herbarium (IN); http://herbarium.infoboss.co.kr/intro.php IB-00567) as a first genome of subgenus Lotos in genus Nymphaea. Total DNA of N. lotus was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea, and de novo assembly was conducted by Velvet 1.2.10 (Zerbino and Birney Citation2008), gap filling was done by SOAPGapCloser 1.12 (Zhao et al. Citation2011), and all bases were confirmed by alignment results generated by BWA 0.7.17, Addison, TX (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation.
The chloroplast genome of N. lotus (Genbank accession is MK040443) is 159,311 bp and has four subregions: 89,610 bp of large single copy (LSC) and 19,333 bp of small single copy (SSC) regions are separated by 25,184 bp of inverted repeat (IR). It contained 130 genes (85 protein-coding genes, 8 rRNAs, and 37 tRNAs); 17 genes (6 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions. The overall GC content of N. lotus is 39.1% and in the LSC, SSC, and IR regions are 37.7%, 34.2%, and 43.4%, respectively.
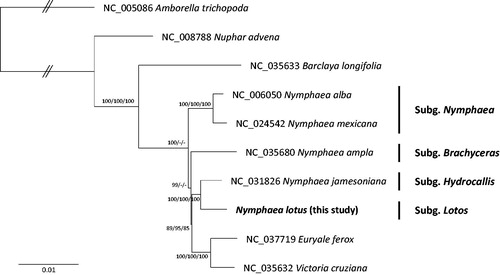
Nine complete Nymphaeaceae chloroplast genomes were used for constructing three phylogenetic trees; neighbor joining and maximum parsimony methods using MEGA X (Kumar et al. Citation2018) and maximum likelihood tree using IQ-TREE 1.6.6 (Nguyen et al. Citation2015). Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013). The phylogenetic tree confirmed that subgenus Lotos was clustered with subgenus Hydrocallis (). However, Euryale ferox and Victoria cruziana were nested in the clade of Nymphaea which is different from the previous study (Borsch et al. Citation2007). It indicates that additional chloroplast genomes will be required for clarifying phylogenetic relationships of Nymphaea and neighbour genera.
Figure 1. Neighbor joining, maximum parsimony, and maximum likelihood phylogenetic trees (bootstrap repeat is 10,000 for the three trees) of nine Nymphaeaceae chloroplast genomes and one outgroup species: Nymphea lotus (MK040443, this study), Nymphaea ampla (NC_035680), Nymphaea jamesoniana (NC_031826), Nymphaea mexicana (NC_024542), Nymphaea alba (NC_006050), Euryale ferox (NC_037719), Barclaya longifolia (NC_035633), Nuphar advena (NC_008788), Victoria cruziana (NC_035632), and Amborella trichopoda (NC_005086). The numbers above branches indicate bootstrap support values of maximum likelihood, neighbor joining tree, and maximum parsimony tree, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Albert VA, Barbazuk WB, Der JP, Leebens-Mack J, Ma H, Palmer JD, Rounsley S, Sankoff D, Schuster SC, Soltis DE. 2013. The Amborella genome and the evolution of flowering plants. Science. 342:1241089.
- Borsch T, Hilu KW, Wiersema JH, Löhne C, Barthlott W, Wilde V. 2007. Phylogeny of Nymphaea (Nymphaeaceae): evidence from substitutions and microstructural changes in the chloroplast trnT-trnF region. Int J Plant Sci. 168:639–671.
- Chen F, Liu X, Yu C, Chen Y, Tang H, Zhang L. 2017. Water lilies as emerging models for Darwin’s abominable mystery. Hortic Res. 4:17051.
- Conard HS. 1905. The water lilies: a monograph of the genus Nymphaea. Washington (DC): Carnegie Institution.
- Hirthe G, Porembski S. 2003. Pollination of Nymphaea lotus (Nymphaeaceae) by rhinoceros beetles and bees in the northeastern Ivory Coast. Plant Biol. 5:670–676.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
