Abstract
We report the complete mitogenome of Draco maculatus, which is 16,708 bp in size and includes 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a control region. PCGs, with 13 genes, is 11,280 bp in length. All PCGs use ATN as the start codon; COI, ATP8, ND4L, ND5, and Cytb use the typical stop codon TAA; but ATP6, COIII, and ND3 stop with a single T. Phylogeny reconstructed using the Bayesian inference (BI) method with 13 PCGs indicates that D. maculatus is located at the root of Draconinae.
Spotted flying dragon (Draco maculatus), with a capacity of gliding among trees, is an endemic agamid lizard in Southeast Asia from Assam and Yunnan to Singapore (Zhao and Adler Citation1993). Phylogeny reconstructed according to mitogenomic data would indicate the evolutionary history of agamid lizards. So far, mitogenomic data of agamid lizards are only available for 27 species (including 15, 4, 4, 2, 1 and 1 species in Agaminae, Leiolepidinae, Draconinae, Amphibolurinae, Hydrosaurinae and Uromastycinae, respectively) of the more than 480 species (Jin and Brown Citation2018; Yu et al. Citation2018). However, the phylogenetic position and inter-relationships of the subfamilies are complex and controversial (Macey et al. Citation2008; Okajima and Kumazawa Citation2010; Pyron et al. Citation2013). In order to obtain more basic genetic information on this group of lizards, we determined the complete mitogenome of D. maculatus.
The sample (voucher number DYHZ301) was collected from Hainan, China, and stored at −80 °C in our laboratory at Hangzhou Normal University. Total genomic DNA was extracted by the Qiagen Genomic DNA Kit (Hilden, Germany). The whole mitochondrial genome was amplified with the highly conserved primers (seven pairs) and the species-specific primers (24 pairs). The resultant reads were assembled and annotated using MEGA X (Kumer et al. Citation2018). We conducted the analyses following a study on another agamid lizard (Yu et al. Citation2018).
The complete and circular mitochondrial genome is 16,708 bp in size (GenBank Accession No. KY073263), with an AT bias of 55.2%, and includes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (12S rRNA and 16S rRNA), and a non-coding AT-rich region (control region or D-loop). There are one ND6 subunit gene and seven tRNA on the L-strand, and the other 28 genes in D. maculatus on the H-strand. Thirteen PCGs are 11,280 bp in size and start with typical ATN codons; the TAA was found as the stop codon in CO1, ATP8, ND4L, ND5 and Cytb; and the TAG was found as the stop codon in ND1, ND2, COII, ND4 and ND6; while 3 PCGs (ATP6, COIII and ND3) stop with a single T. The 22 tRNA genes varied in size from 57 bp (tRNACys) to 73 bp (tRNAAsn). The overall base composition of D-loop (1563 bp) is A (32.9%), T (34.8%), C (20.0%) and G (12.3%). The longest intergenic region (28 bp) is located between COI and tRNASer(UCN).
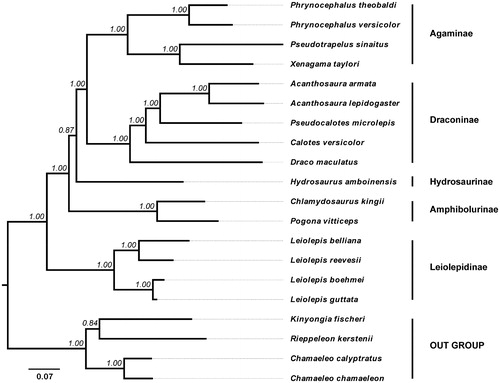
Selecting four Chamaeleonid species as outgroups, the phylogeny of 16 Agamid species including D. maculatus was reconstructed based on 13 PCGs by the Bayesian inference (BI) methods through MrBayes (version 3.2.2). The resultant BI tree distinctly indicated that D. maculatus belongs to a monophyletic group including Acanthosaura, Pseudocalotes, and Calotes, and is located at the root of the subfamily Draconinae ().
Disclosure statement
The authors report no conflict of interest. All the authors approved the final version of the manuscript for publication and agreed to be held accountable for the content of this manuscript.
Additional information
Funding
References
- Jin YT, Brown RP. 2018. Partition number, rate priors and unreliable divergence times in Bayesian phylogenetic dating. Cladistics. 34:568–573.
- Kumer S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Macey JR, Kuehl JV, Larson A, Robinson MD, Ugurtas IH, Ananjeva NB, Rahman H, Javed HI, Osman RM, Doumma A, Papenfuss TJ. 2008. Socotra Island the forgotten fragment of Gondwana: unmasking chameleon lizard history with complete mitochondrial genomic data. Mol Phylogenet Evol. 49:1015–1018.
- Okajima Y, Kumazawa Y. 2010. Mitochondrial genomes of acrodont lizards: timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol Biol. 10:1–15.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:635–653.
- Yu XL, Du Y, Fang MC, Li H, Lin LH. 2018. The mitochondrial genome of Pseudocalotes microlepis (Squamata: Agamidae) and its phylogenetic position in Agamids. Asian Herprtol Res. 9:24–34.
- Zhao EM, Adler K. 1993. Herpetology of China. Oxford, Ohio: SSAR.