Abstract
The complete mitochondrial genome of the orb-weaving spider Neoscona scylla was characterized in this study. The circular molecule is 14,092 bp in length (GenBank accession no. MK086023), containing 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a control region. The nucleotide composition is asymmetric (35.27% A, 9.82% C, 15.55% G, 39.36% T), with an overall A + T content of 74.63%. The gene arrangement of N. scylla is identical to that observed in other spiders. Nine PCGs are initiated with typical ATN start codons, three genes (cox2, cox3, and nad6) begin with TTG, and cox1 use TTA as initiation codon. Twelve PCGs stop with complete termination codon TAA and TAG, while nad3 uses incomplete termination codon (T). Twenty-five reading frame overlaps and five intergenic regions are found in the mitogenome of N. scylla. Seven tRNAs lose TΨC arm stems, whereas three lack the dihydrouracil (DHU) arm. The phylogenetic relationships based on 13 PCGs show that N. scylla clusters closest to 5 species of Araneidae.
Neoscona scylla is a type of orb-weaving spider belonging to the order Araneae and family Araneidae (Platnick Citation2015). The species is widely distributed in China, Russia and Japan. N. scylla captures insect pests as food through weaving horizontal or upright web, and is recognized as an important natural enemy for biological control (Wang et al. Citation1996; Li et al. Citation2014). In this study, samples of N. scylla were collected from the paddy fields in Libo county (N25 °19′, E107 °56′), Guizhou Province, China, and stored in the spider specimen room of Guiyang University with an accession number GYU-GZML-10.
The complete mitogenome of N. scylla is a circular molecule of 14,092 bp in length (GenBank under accession no. MK086023), and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rrnL and rrnS), and a non-coding region (putative control region). The gene organization of N. scylla is similar to that observed in other spiders (Wang et al. Citation2014; Li et al. Citation2016). The nucleotide composition of N. scylla mitogenome is asymmetric (35.27% A, 9.82% C, 15.55% G, 39.36% T), with an overall A + T content of 74.63%. The AT-skew and GC-skew of this genome were −0.055 and 0.226, respectively. Twenty-two genes were encoded on the major strand (J-strand), whereas the others were encoded on the minor strand (N-strand).
The N. scylla mitogenome harbours a total of 23 bp intergenic spacer sequences, which is made up of five regions in the range from 3 to 7 bp. Gene overlaps were found at 25 gene junctions and involved a total of 221 bp, the longest 28 bp overlapping located between nad3 and trnL2. Among 22 tRNAs, 10 of them appear to be truncated and lack the potential to form the cloverleaf-shaped secondary structure. Seven tRNAs (trnW, trnK, trnE, trnF, trnH, trnT, and trnL1) lost the TΨC-stem, three tRNAs (trnA, trnS1, and trnS2) lost the dihydrouracil (DHU) arm. The length of these tRNAs ranged from 50 bp (trnC) to 67 bp (trnM and trnD), A + T content ranged from 67.69% (trnR) to 84.75% (trnG). The rrnL was located between trnL1 and trnV, while the rrnS resided between trnV and trnQ. The rrnL was 1022 bp in length with A + T content of 78.67%, and rrnS was 694 bp in length with A + T content of 77.81%. The control region was located between trnQ and trnM genes with a length of 508 bp, and the A + T content was 78.54%.
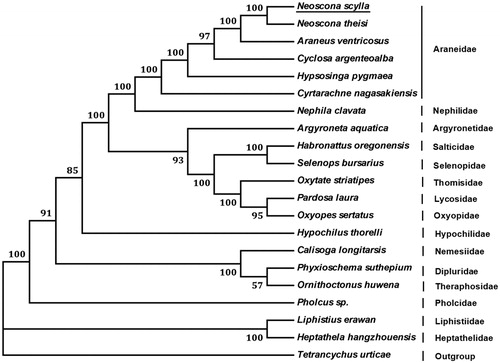
Nine PCGs in N. scylla mitogenome start with a typical ATN (ATA, ATC, and ATT) codon, three genes (cox2, cox3, and nad6) begin with TTG, and cox1 uses TTA as initiation codon. Twelve PCGs stop with complete termination codon (TAA and TAG), while nad3 uses incomplete codon (T) as termination codon. We constructed the phylogenetic relationships of N. scylla and 19 other spiders based on the 13 PCGs amino acids using neighbour-joining method. The results have shown that N. scylla is closely clustered with other five spiders in family Araneidae (), which agree with the morphology classifications.
Figure 1. Phylogenetic tree showing the relationship between N. scylla and 19 other spiders based on neighbour-joining method. Tetrancychus urticae was used as an outgroup. GenBank accession numbers used in the study are the following: Neoscona theisi (NC_026290.1), Araneus ventricosus (KM588668.1), Hypsosinga pygmaea (KR259803.1), Cyrtarachne nagasakiensis (KR259802.1), Cyclosa argenteoalba (KP862583.1), Nephila clavata (NC_008063.1), Argyroneta aquatica (NC_026863.1), Habronattus oregonensis (AY571145.1), Selenops bursarius (NC_024878.1), Oxytate striatipes (NC_025557.1), Pardosa laura (KM272948.1), Oxyopes sertatus (NC_025224.1), Pholcus sp. (KJ782458.1), Phyxioschema suthepium (NC_020322.1), Ornithoctonus huwena (AY309259.1), Calisoga longitarsis (EU523754.1), Hypochilus thorelli (EU523753.1), Liphistius erawan (NC_020323.1), Heptathela hangzhouensis (AY309258.1), and T. urticae (EU345430.1). Spider determined in this study was underlined.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Li C, Wang ZL, Fang WY, Yu XP. 2016. The complete mitochondrial genomes of two orb-weaving spider Cyrtarachne nagasakiensis (Strand, 1918) and Hypsosinga pygmaea (Sundevall, 1831) (Araneae: Araneidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2811–2812.
- Li QJ, Wang J, Wei BY, Wang Z. 2014. Bio-ecology of the Neoscona scylla. Acta Laser Biol Sin. 42:61–164.
- Platnick NI. 2015. The world spider catalog, version 13.5. American museum of natural history. http://research.amnh.org/iz/spides/catalog/.
- Wang HQ, Yan HM, Yang HM. 1996. Studies on the ecology of spiders in paddy fields and utilization of spiders for biological control in China. Sci Agr Sin. 29:68–75.
- Wang ZL, Li C, Fang WY, Yu XP. 2014. The complete mitochondrial genome of wolf spider Pirata subpiraticus Boes.et str. (Araneae: Lycosidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1926–1927.
