Abstract
The complete mitochondrial genome of the Southeast Asian shrew (Crocidura fuliginosa) was first sequenced and characterized. The genome was 16,967 base pairs (bp) in length containing 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and a control region. The composition and arrangement of genes were analogous to other vertebrates. All PCGs were used to construct phylogenetic tree for C. fuliginosa and other 12 Crocidura species available on GenBank. Phylogenetic result indicated that C. fuliginosa has a close relationship with C. lasiura and C. attenuate. Our results will provide information for further molecular studies about this genus.
White-toothed shrews, Genus Crocidura, contains 172 species, among them 11 species are distributed in China (Smith et al. Citation2013). Crocidura fuliginosa is the largest white-toothed shrew in China and occurs in Southwestern and Eastern China (Jiang and Hoffmann Citation2001). Although there are many studies on C. fuliginosa, its taxon and distribution have been controversial (Hutterer Citation2005; Dubey et al. Citation2008; Bannikova et al. Citation2011; Abramov et al. Citation2012). In order to provide more molecular information for this species, we first sequenced and characterized the complete mitochondrial genome of C. fuliginosa.
The voucher specimen (S0687) was collected from Xinping of Sichuan province (24.216°N, 101.491°E), China and stored in Shandong University (Weihai). Total genomic DNA was isolated from muscle tissue fixed in 95% ethanol using the EasyPure Genomic DNA kit (TransGen Biotech Co., Ltd, Beijing, China). Whole mitochondrial genome was amplified with 18 pairs of primers and sequenced by protocols that are routinely used in our laboratory (Wang et al. 2016). The resultant sequences were assembled and annotated using the BioEdit 7.2.5 (Hall Citation1999) and MEGA 7.0.14 software (Kumar et al. Citation2016).
The complete mitogenome of C. fuliginosa was 16,967 bp in length and has been deposited in GenBank under the accession number MK032069. It contains 37 genes arranged in the same order with other reported vertebrate mitogenomes, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region. Most genes are encoded on the heavy (H) strand, except for the nicotinamide adenine dinucleotide dehydrogenase subunit (ND) 6 and 8 tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) which are encoded on the light (L) strand. All PCGs start with an ATG codon except ND2 and DN5 start with CGA. Cytb ends with AGA and other PCGs end with TAA, TA, and T. All tRNA genes arranged from 59 to 75 bp and can be folded into typical clover secondary structure except for tRNA-Ser (AGY) in which its dihydrouridine (DHU) arm is lacking. The control region is 1506 bp in length and located between tRNA-Pro and tRNA-Phe genes. The overall base composition is 32.5% for A, 31.7% for T, 13.3% for G, and 22.5% for C.
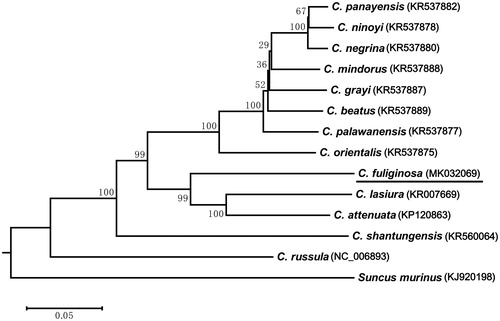
To investigate relationships between C. fuliginosa and other Crocidura species, we constructed the phylogenetic tree based on 13 PCGs by the ML method using RAxML v7 (Stamatakis Citation2006) including other 12 Crocidura species available on GenBank, and Suncus murinus was used as outgroup. The result showed that C. fuliginosa has a sister relationship with C. lasiura and C. attenuate (). The complete mitochondrial genome of C. fuliginosa and phylogenetic analyses will shed light on evolutionary relationship within species in genus Crocidura and will be useful for species delimitation, phylogenetic analysis and other relative studies.
Acknowledgments
The authors thank the anonymous reviewers for providing valuable comments on this article.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Abramov AV, Bannikova AA, Rozhnov VV. 2012. White-toothed shrews (Mammalia, Soricomorpha, Crocidura) of coastal islands of Vietnam. Zookeys. 207:37–47.
- Bannikova AA, Abramov AV, Borisenko AV, Lebedev VS, Rozhnov VV. 2011. Mitochondrial diversity of the white-toothed shrews (Mammalia, Eulipotyphla, Crocidura) in Vietnam. Zootaxa. 2812:1–20.
- Dubey S, Salamin N, Ruedi M, Barriere P, Colyn M, Vogel P. 2008. Biogeographic origin and radiation of the Old World crocidurine shrews (Mammalia: Soricidae) inferred from mitochondrial and nuclear genes. Mol Phylogenet Evol. 48:953–963.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Hutterer R. 2005. Order Soricomorpha. In: Wilson DE, Reeder DM, editors. Mammal species of the world. A taxonomic and geographic reference. 3rd ed. Baltimore: Johns Hopkins University Press. p. 220–311.
- Jiang XL, Hoffmann RS. 2001. A revision of the white-toothed shrews (Crocidura) of southern China. J Mammal. 82:1059–1079.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Smith AT, Xie Y, Hoffmann RS, Lunde D, MacKinnon J, Wilson DE, Wozencraft WC. 2013. Mammals of China. Princeton (NJ): Princeton University Press.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wang SH, Cong HY, Kong LM, Motokawa M, Li YC. 2016. Complete mitochondrial genome of the greater bandicoot rat Bandicota indica (Rodentia: Muridae). Mitochondrial DNA. 27:4349–4350.