Abstract
Misgurnus anguillicaudatus is a highly valued, aquaculture-relevant food fish in East Asian countries. Because of overfishing and environmental pollution, the number of wild Nansi loach has been sharply decreased in these years. In this study, the complete mitochondrial genome of Nansi loach was obtained by PCR. The genome is 16,646 bp in length, including 2 ribosomal RNA genes. 13 proteins-coding genes, 22 transfer RNA genes, and a non-coding control region, the gene composition and order of which was similar to most reported from other vertebrates. Sequence analysis showed that the overall base composition is 29.8% for A, 28.2% for T, 25.6% for C, and 16.4% for G. The phylogenetic tree showed that Misgurnus family got together for one branch, which includes Nansi M. anguillicaudatus, and the other loaches had their own branches. Also, the mitochondrial genome sequence of M. anguillicaudatus was aligned by BLAST, compared with Cobitinae the sequence similarity could reach >95%, and the similarity to Misgurnus was >99%.
Cyprinid loach, Misgurnus anguillicaudatus (Cypriniformes; Cobitidae), a small-sized freshwater fish species, is a highly valued, aquaculture-relevant food fish in East Asian countries. This loach species has also been given much attention as an important model organism to study developmental biology, polyploidy evolution, and genetic breeding (Liang et al. Citation2018; Zhang et al. Citation2018). Because of overfishing and environmental pollution of Nansi Lake, the number of natural wild M. anguillicaudatus has been sharply decreased in these years. Therefore, it is very important to characterize the complete mitogenome of this species, which could be a fundamental basis to address genetic identity and diversity in future conservation program of this rarely occurring (Sang et al. Citation2017).
In this study, we sequenced the complete mitogenome of M. anguillicaudatus with a GenBank accession number MK093946. The voucher specimen was collected from Nansi Lake, north latitude 34°66″ and east longitude 117°20″, Jining city, China. They were preserved in 95% alcohol, which were stored in biology herbarium of Heze University. All DNA were extracted using phenol–chloroform extraction methods and stored at −80 °C. The mitogenome was amplified by primers which were initially published (Zeng et al. Citation2012). The entire mitogenome sequence of Nansi loach was 16,646 bp in length, consisting of 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, one replication origin (OL), and one control region (D-loop). From the base composition analysis, the percent A + T content was 58.0% (29.8% for A, 28.2% for T, 25.6% for C, and 16.4% for G). Twelve PCGs, 14 tRNA genes, and two rRNA genes were located on the heavy strand (H-strand), while one PCG (ND6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro) on the light strand (L strand). Eight PCGs (ND1, COI, COII, ATP8, ATP6, ND4L, ND5, and ND6) were terminated with TAA stop codon and three PCGs (ND2, ND3, and ND4) ended with TAG. On the other hand, remaining two PCGs (COIII and CYTB) ended with the incomplete stop codon represented as a single T. Frame overlapping occurred at three pairs of PCGs. ATP8 and ATP6 overlapped by 10 nucleotides (nt), ND4L and ND4 by 7 nt, and ND5 and ND6 (encoded on opposing stand) by 4 nt. Two ribosomal RNA genes, 12S rRNA (953 bp) and 16S rRNA (1679 bp), were located between tRNAPhe and tRNALeu with a separation by the tRNAVal as seen in other vertebrate mitogenomes.
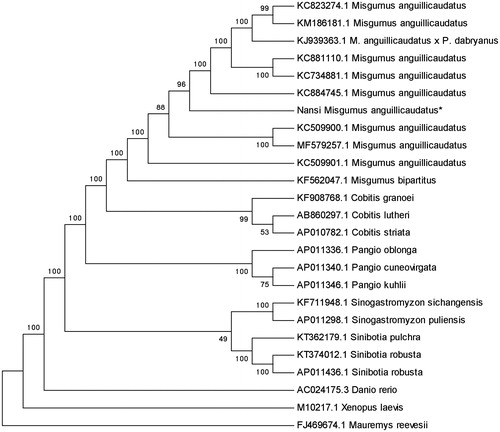
To determine taxonomic status of M. anguillicaudatus, we performed the phylogenetic relationship of Nansi M. anguillicaudatus stock with other natural populations in loach as inferred by entire mitogenome (Liang et al. Citation2018). The phylogenetic tree showed that Misgurnus family got together for one branch, which includes Nansi M. anguillicaudatus, and the other loaches had their own branches (). Also, the mitochondrial genome sequence of M. anguillicaudatus was aligned by BLAST, compared with Cobitinae the sequence similarity could reach >95%, and the similarity to Misgurnus was >99%.
Figure 1. Phylogenetic relationship of Nansi M. anguillicaudatus stock with other loach as inferred by entire mitogenome. *The Nansi loach (accession number: MK093946) in the position of the evolutionary tree. Trees were reconstructed using MEGA 7 program (Kumar, Tamura, Nei) with neighbor-joining method. Numbers above branches are bootstrap values by 1000 replicates. The phylogenetic tree showed that Nansi loach to be one of Misgurnus, and the other loaches had their own branches.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Liang X, Zhu D, Li X, Cai K, Zhang H, Zhang G. 2018. Complete mitochondrial genome of the hybrid loach of Paramisgurnus dabryanus ssp. (female) and Misgurnus anguillicaudatus (male). Mitochondrial DNA Part B. 3:336–337.
- Sang YL, Bang IC, Nam YK. 2017. Complete mitochondrial genome of albino cyprinid loach, Misgurnus anguillicaudatus, (Cypriniformes: Cobitidae). Conserv Genetics Resour. 1:1–4.
- Zeng L, Wang J, Sheng J, Gu Q, Hong Y. 2012. Molecular characteristics of mitochondrial DNA and phylogenetic analysis of the loach (Misgurnus anguillicaudatus) from the poyang lake. Mitochondrial DNA. 23:187–200.
- Zhang G, Zhu D, Li X, Liang X, Cai K, Zhang H, Zhang G. 2018. *. Complete mitochondrial genome of natural diploid loaches Misgurnus anguillicaudatus from the Taihu Lake. Mitochondrial DNA Part B. 3:566–567.
