Abstract
Paeonia jishanensis is endemic in China, and has been proven to be one of the most important ancestral species of cultivated tree peonies. Wild P. jishanensis has been over-exploited and becomes quite rare in China. The wild peony of P. jishanensis was used as an experimental material to sequence the chloroplast genome (cpDNA) by high-throughput sequencing (NCBI accession number: MG991935). The cpDNA of P. jishanensis is a typical quadripartite structure with a length of 152,628 bp, including a large single-copy (LSC) region of 84,292 bp and a small single-copy (SSC) region of 17,044 bp separated by a pair of identical inverted repeat regions (IRs) of 25,646 bp each. The cpDNA contains 112 genes, including 78 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes. The result of phylogenomic analysis supports the difference of tree peony from the herbaceous peony.
Tree peonies refer to plants belonging to genus Paeonia sect. Moutan DC. (APG IV Citation2016). Besides its reputation as ‘the king of flowers’ and ornamental use, peonies are also of high economic value. Its root bark, referred to as ‘peony bark,’ is a common medicinal material in compound traditional Chinese medicine prescriptions and is listed in the Chinese pharmacopeia. Peony seed oil is rich in alpha-linolenic acid and other unsaturated fatty acids and is a healthy edible oil with promising future applications (Hong et al. Citation2017). Peonies are cultivated in various places around the world. China’s unique wild peonies have always been regarded as a valuable germplasm resource locally and abroad. As one of the seven wild peonies, Paeonia jishanensis is regarded as an important ancestral species (Xu et al. Citation2016). The present study has assembled and analyzed the chloroplast genome of P. jishanensis as a major Paeonia germplasm resource, as well as provided foundational data for phylogenetics research.
Fresh leaves of P. jishanensis were collected from Majiagou in Jishan County, Shanxi Province, China (35°43′N, 110°58′E). The herbarium vouchers of the plants used in this study are deposited in the Luoyang Normal University Specimen Museum under accession number PLANT20171101. Genomic DNA was extracted from the leaves using Plant DNA extraction kit (Tiangen, Beijing). DNA samples that passed the quality test were sonicated into fragments. After purification, end-terminal repair, 3′ A addition, and adapter ligation were conducted. Agarose gel electrophoresis was used to select fragments sizes. Sequencing libraries were constructed after PCR amplification and were sequenced using the Illumina HiSeq 2500 platform (Illumina, Hayward, CA). Raw data obtained from sequencing were filtered using the CLC 9.0 software to remove low-quality sequences (Wambugu et al. Citation2015). The chloroplast genome from Paeonia ludlowii was used as a reference (Li et al. Citation2018). The complete chloroplast genome of P. jishanensis was annotated in Geneious R9 (v9.0.2) (Wang et al. Citation2017) and then submitted to GenBank (accession no. MG991935).
The entire length of the sequence is 152,628 bp, with a GC content of 38.4%. The LSC region is 84,292 bp in length, the SSC region is 17,044 bp, and the IR region is 25,646 bp in length. The GC content of the LSC, SSC, and IR regions in P. jishanensis are 36.7%, 32.7%, and 43.1%, respectively. Chloroplast annotation indicated that the P. jishanensis chloroplast genome contains a total of 112 genes, including 78 protein-encoding genes, 30 tRNA genes, and 4 rRNA genes. Among these genes, 19 genes are duplicated within the IR region. The LSC region contains 22 tRNA genes and 60 protein-encoding genes, whereas the SSC region contains 1 tRNA gene and 10 protein-encoding genes. Among the protein-encoding genes in P. jishanensis, nine genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16) contain one intron and three genes (clpP, rps12, ycf3) have two introns.
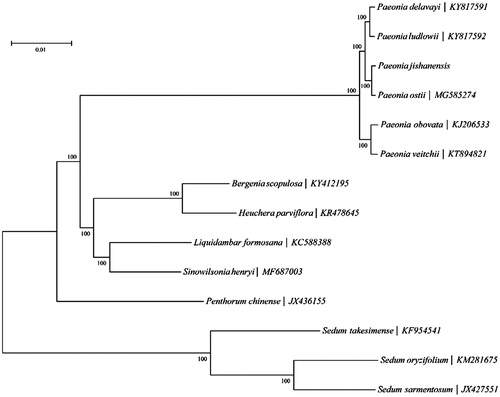
Thirteen whole chloroplast genomes from the order Saxifragales were downloaded from GenBank and used in multiple sequence alignment with the chloroplast genome of P. jishanensis (). The MEGA6 software (Tamura et al. Citation2013) was used to construct neighbour-joining phylogenetic trees (Bootstrap value =1000). The results showed that four kinds of tree peony (P. jishanensis, P. ostii, P. ludlowii and P. delavayi) were situated within the same branch. Two kinds of herbaceous peony, P. veitchii and P. obovata cluster together.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot J Linn Soc. 181:1–20.
- Hong DY, Zhou SL, He XJ, Yuan JH, Zhang YL, Cheng FY, Zeng XL, Wang Y, Zhang XX. 2017. Current status of wild tree peony species with special reference to conservation. Biodiversity Sci. 25:781–793.
- Li H, Guo Q, Zheng W. 2018. Characterization of the complete chloroplast genomes of two sister species of Paeonia: genome structure and evolution. Conservation Genet Resour. 10:209–212.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wambugu PW, Brozynska M, Furtado A. 2015. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci Rep. 5:13957.
- Wang XF, Yang N, Su J, Zhang H, Cao XY. 2017. The complete chloroplast genome of Gentiana macrophylla. Mitochondrial DNA Part B. 2:395–396.
- Xu XX, Cheng FY, Xian HL, Peng LP. 2016. Genetic diversity and population structure of endangered endemic Paeonia jishanensis in China and conservation implications. Biochem Syst Ecol. 66:319–325.