Abstract
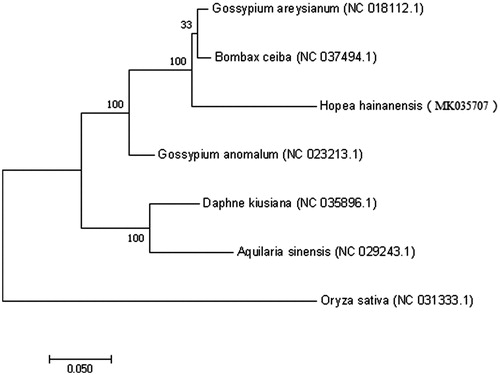
Hopea hainanensis is an endangered species with scattered occurrence, and it is one of the representative species of tropical rainforest in Hainan Island. In this paper, we reported and characterized the complete chloroplast genome sequence of the species assembled from short reads generated by Illumina sequencing. Total chloroplast genome size was 151,795 bp, including inverted repeats (IRs) of 26,098 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 84,419 bp and 19,520 bp, respectively. A total of 136 genes, including 45 tRNA, 8 rRNA, and 83 protein-coding genes, were identified. The GC content of H. hainanensis is 37.3%. Phylogenetic analysis using the maximum likelihood algorithm showed that H. hainanensis is sister to a clade with five species in the order Dicotyledoneae.
Hopea hainanensis Merr. et Chun (Dipterocarpaceae) is an endangered species with scattered occurrence, and it is one of the representative species of the tropical rainforest, as well as an important timber species and aromatic plants with high ornamental and economic value (Kammesheidt et al. Citation2003). The overexploitation of the species has led to the drastic reduction of their population (Lan et al. Citation2007). In recent years, research on H. hainanensis has focussed on its geographical distributions, seed characteristics and chemical compounds (Chen et al. Citation2015; Xu et al. Citation2018). The information of chloroplast genomes has been extensively applied to understanding plant genetic diversity and conservation genetics, while studies regarding this plant are lacking (Cvetković et al. Citation2017). In this paper, we characterized the complete chloroplast genome sequence of H. hainanensis to contribute for further phylogenetical and protective studies of this plant.
Fresh leaves were collected from five individuals of H. hainanensis in BaWang Ling, Hainan Island (N19°04'51.39", E109°07'22.83"), China. The specimen was deposited at the botany laboratory of Hainan Normal University, Haikou, China. The total genomic DNA was extracted from ten mixed fresh leaves of H. hainanensis using the modified CTAB method (Doyle Citation1987). Genome sequencing was performed on an Illumina Hiseq X Ten platform with paired-end reads of 150 bp. In total, 76.4 Mb short sequence data with Q20 was 97.26% was obtained. Low-quality reads were filtered out and the remaining high-quality reads were used to assemble the chloroplast genome in SOAPdenovo (Luo et al. Citation2012). The genes annotation in the chloroplast genome was carried out with the DOGMA Programme (Wyman et al. Citation2004). The circular chloroplast genome map was drawn by using OGDRAW and the phylogenomic tree was constructed using MAFFT (Katoh and Standley Citation2013). The complete chloroplast genome sequence was submitted to Genbank under the accession number is MK035707.
Seven chloroplast genome sequences were aligned using the complete chloroplast genomes including H. hainanensis, and one species from Gramineae (Oryza sativa) was used as the outgroup species. Phylogenetic analysis using the maximum likelihood algorithm was conducted with RAxML (Stamatakis Citation2014) implemented in Geneious ver. 10.1(Kearse et al. Citation2012).
A typical quadripartite structure was found in the chloroplast genome of H. hainanensis with a length of 151,795 bp, which contains inverted repeats (IRs) of 26,098 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 84,419 bp and 19,520 bp, respectively. The chloroplast genome has a GC content of 37.3% and contains a total of 136 predicted genes, including 45 tRNAs, 8 rRNAs and 83 protein-coding genes. Among them, six protein-coding genes (rpoC1, rps16, petD, rpl2, nadhA and nadhB) contain two introns and two genes (rps12 and ycf3) contain three introns. Phylogenetic analysis with other six plant chloroplast genomes showed that H. hainanensis is sister to a clade containing five species in Dicotyledoneae (). The Hopea hainanensis chloroplast genome provides essential and important DNA molecular data for phylogenetic and evolutionary analysis for this endangered species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen HX, Huang CT, He F, Zheng W, Feng JP. 2015. Review on research progress of Hopea hainanensis. Trop Forestry. 43:4–6.
- Cvetković T, Hinsinger DD, Strijk JS. 2017. The first complete chloroplast sequence of a major tropical timber tree in the meranti family: vatica odorata (dipterocarpaceae). Mitochondrial DNA. 2:52–53.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kammesheidt L, Dagang AA, Schwarzwäller W, Weidelt HJ. 2003. Growth patterns of dipterocarps in treated and untreated plots. Forest Ecol Manage. 174:437–445.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lan QY, Jiang XC, Song SQ, Lei YB, Yin SH. 2007. Changes in germinability and desiccation-sensitivity of recalcitrant Hopea hainanensis Merr. et Chun seeds during development. Seed Sci Technol. 35:21–31.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Xu WJ, Liang Y, Fu CY, Chen GB. 2018. Chemical compounds from volatile oil of Hopea hainanensis. J Chin Med Mater. 41:1872–1874.