Abstract
Prunus speciosa (Oshima cherry) is a flowering tree species with high ornamental and economic values and is also an indigenous species with a very limited natural distribution in Japan. Here we determined the first complete chloroplast genome of P. speciosa using genome skimming approach. The cp genome was 157,916 bp long, with a large single-copy region (LSC) of 85,927 bp and a small single-copy region (SSC) of 19,123 bp separated by a pair of inverted repeats (IRs) of 26,433 bp. It encodes 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes. The phylogenetic analysis indicated that P. speciosa is close related with ‘P. serrulata var. spontanea’ auct. non. (P. leveilleana).
Prunus speciosa (Koidz.) Nakai (Oshima cherry, Rosaceae), also known as Cerasus speciosa (Koidz.) H. Ohba, is a flowering tree species with high ornamental and economic values (Wilson Citation1916; Ohba Citation2001). Prunus speciosa was important in originating many of the garden cultivars, especially in many fragrant cultivars (Ohba et al. Citation2007). Prunus speciosa is indigenous on Oshima island and other neighboring Izu Islands, and also on Izu Peninsula and Boso Peninsula in Honshu island. But P. speciosa on the peninsulas were thought to be brought to Honshu long ago for the usage of firewood, and recent analyzing shows an introgressive hybridization with Prunus jamasakura in the peninsula populations (Kuitert and Peterse Citation1999; Katsuki Citation2015). According to recent researches, P. speciosa was clearly distinguished from its related taxa both genetically and morphologically (Chang et al. Citation2007; Kato et al. Citation2014; Katsuki and Iketani Citation2016), but the genetic relationship of P. speciosa relative to other flowering cherries has not been well established.
Total genomic DNA was extracted from silica-dried leaves collected from the nursery (Ind. No. IZO-2014-06) in Tama Forest Science Garden (Hachioji, Japan) using a modified CTAB method (Doyle and Doyle Citation1987). The P. speciosa tree in the nursery was introduced from natural populations on Oshima island. A voucher specimen (Sun1704123) was collected and deposited in the Herbarium of Zhejiang Academy of Forestry (HZJAF). DNA libraries preparation and pair-end 125 bp read length sequencing were obtained on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA). About 11.8 Gb of raw data were trimmed and assembled into contigs using CLC Genomics Workbench 8. Then, all the contigs were aligned to the reference cp genome of Prunus takesimensis (MG754959; Cho et al. Citation2018) using BLAST (NCBI BLAST v2.2.31) search and the draft cp genome of P. speciosa was constructed by connecting overlapping terminal sequences in Geneious R11 software (Biomatters Ltd., Auckland, New Zealand). Gene annotation was performed via the online program Dual Organellar Genome Annotator (DOGMA; Wyman et al. Citation2004).
The complete cp genome of P. speciosa (GenBank accession MH998233) was 157,916 bp long consisting of a pair of inverted repeat regions (IRs with 26,433 bp) divided by two single-copy regions (LSC with 85,927 bp; SSC with 19,123 bp). The overall GC content of the total length, LSC, SSC, and IR regions was 36.7%, 34.6%, 30.2%, and 42.5%, respectively. The whole cp genome encoded 129 genes including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
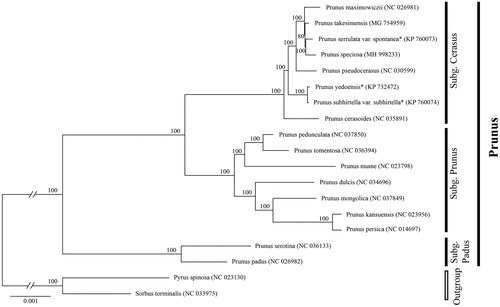
Phylogenetic analysis was conducted to confirm the phylogenetic position of newly sequenced P. speciosa amongst 17 representative Prunus L. species and two outgroup taxa. We reconstructed a phylogeny employing the GTR + R + I model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference in RAxML-HPC v.8.2.10 on the CIPRES cluster (Miller et al. Citation2010). The ML tree () was consistent with the most recent phylogenetic study on Prunus (Shi et al. Citation2013; Chin et al. Citation2014). Illumina, San Diego, CA speciosa belongs to subg. Cerasus as expected and exhibited the closest relationship with ‘P. serrulata var. spontanea’ auct. non. Maxim. (P. leveilleana Koehne).
Disclosure statement
The authors are really grateful to the opened raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chang KS, Chang CS, Park TY, Roh MS. 2007. Reconsideration of the Prunus serrulata complex (Rosaceae) and related taxa in eastern Asia. Bot J Linn Soc. 154:35–54.
- Chin SW, Shaw J, Haberle R, Wen J, Potter D. 2014. Diversification of almonds, peaches, plums and cherries – molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogenet Evol. 76:34–48.
- Cho MS, Yang JY, Kim SC. 2018. Complete chloroplast genome of Ulleung Island endemic flowering cherry, Prunus takesimensis (Rosaceae), in Korea. Mitochondrial DNA B. 3:274–275.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop; Nov 14; New Orleans. p. 1–8.
- Ohba H. 2001. Cerasus Mill. In: Iwatsuki K, Boufford DE, Ohba H, editors. Flora of Japan. Tokyo: Kodansha.
- Ohba H, Kawasaki T, Tanaka H, Kihara H. 2007. Flowering cherries of Japan, new edition. Tokyo: Yama-kei Publishers (Japanese).
- Kato S, Matsumoto A, Yoshimura K, Katsuki T, Iwamoto K, Kawahara T, Mukai Y, Tsuda Y, Ishio S, Nakamura K, et al. 2014. Origins of Japanese flowering cherry (Prunus subgenus Cerasus) cultivars revealed using nuclear SSR markers. Tree Genet Genomes. 10:477–487.
- Katsuki T 2015. Cerasus species in coastal forests of the Sendai Gulf, Tohoku Region, Japan. Japanese J Conserv Ecol. 20:101–103. (Japanese).
- Katsuki T, Iketani H. 2016. Nomenclature of Tokyo cherry (Cerasus × yedoensis ‘somei-yoshino’, Rosaceae) and allied interspecific hybrids based on recent advances in population genetics. Taxon. 65:1415–1419.
- Kuitert W, Peterse A. 1999. Japanese flowering cherries. Portland: Timber Press.
- Shi S, Li J, Sun J, Yu J, Zhou S. 2013. Phylogeny and classification of prunus sensu lato (Rosaceae) ). J Integr Plant Biol. 55:1069–1079.
- Wilson EH. 1916. The cherries of Japan. Cambridge: The University Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.