Abstract
In the study, we report the complete mitochondrial genome of giraffe seahorse, Hippocampus camelopardalis Bianconi, 1854. The genome of H. camelopardalis is found to be 16,523 bp in length, containing 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a control region (D-loop). The overall base composition of H. camelopardalis is 32.46% for A, 23.54% for C, 14.54% for G, and 29.46% for T, respectively. All PCGs use the typical initiation codon ATG, except for COX1 that uses GTG. The lengths of 12S rRNA and 16S rRNA were 939 and 1685 bp, respectively. Phylogenetic analysis results showed that H. camelopardalis is closely related to the Jayakar’s seahorse H. jayakari. The complete mitochondrial genome of Hippocampus camelopardalis provided essential and important molecular data for evolutionary analysis of genus Hippocampus.
Seahorses have been used for thousands of years as important and precious traditional Chinese medicine to treat a range of conditions and ailments in China (Kumaravel et al. Citation2012). The giraffe seahorse Hippocampus camelopardalis Bianconi, 1854 is a demersal and non-migratory seahorse that inhabits estuaries, seagrasses, and reefs. This species has been traded as adulteration of Hippocampus, which was difficult for identification based solely on morphological characteristics (Hou et al. Citation2018). The complete mitochondrial genome is a valuable source of data for phylogenetic analysis and further development for species identification strategies (Cheng et al. Citation2013; Cheng et al. Citation2017a). Here, we determined the complete mitochondrial genome of H. camelopardalis and constructed the phylogenetic relationship among the Syngnathidae species.
The specimen of H. camelopardalis was collected from Chinese material medicine market in Bozhou city of Anhui Province and identified based on the typical morphometric features, such as dark spots on trunk rings and tip of coronet (Lourie et al. Citation2004). The sample of H. camelopardalis (BZ1807-01) was deposited in the collection centre of College of Pharmaceutical Science at Zhejiang Chinese Medical University. The complete mtDNA of H. camelopardalis was amplified by 14 pairs of primers designed according to the published mitochondrial genomes in the Genus Hippocampus (Wang et al. Citation2017; Cheng et al. Citation2017b; Ge et al. Citation2018). The newly-determined sequence of H. camelopardalis with annotated genes was deposited in GenBank under the accession number of MH939459.
The complete mt genome of H. camelopardalis was 16,523 bp in length containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one control region. The contents of nucleotides were 32.46% (A), 23.54 (C), 14.54% (G), and 29.46% (T), with a much higher AT contents (61.92%). Similar with most vertebral mitochondrial genome, nucleotide overlaps and space gaps were very common in H. camelopardalis (Chen et al. Citation2017; Zhu et al. Citation2018). Overall, there were 10 nucleotide overlapping regions totalling 32 bp and eight intergenic spacers totalling 57 bp. All PCGs started with a typical ATG codon, except for COX1 initiated by a GTG start codon. For the stop codon, six genes ended with complete TAA, while the others terminated with a single base T or TA. The lengths of 12S rRNA and 16S rRNA were 939 and 1685 bp, respectively. The 22 tRNA genes ranged from 67 to 76 bp in length. The control region locating between tRNA-Pro and tRNA-Phe was 884 bp in length, ranging from 15,640 to 16,523 bp.
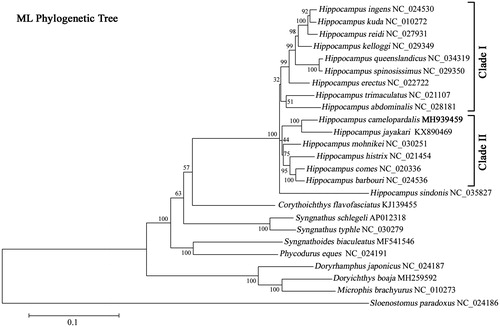
The ML phylogenetic analysis of H. camelopardalis was constructed based on the mitochondrial PCGs using Solenostomus paradoxus as outgroup. H. camelopardalis formed a monophyletic group with H. jayakari with high statistical support, indicating a close genetic relationship between these two species (). Furthermore, the monophyletic group of Hippocampus, which could be divided into three clades, was placed as a sister relationship to Corythoichthys flavofasciatus. The data of H. camelopardalis enriched the resource of Hippocampus genus in systematic and evolutionary biological studies, which would be helpful for researches on genetic diversity and species identification of seahorse.
Figure 1 Maximum-likelihood (ML) phylogenetic tree of Hippocampus camelopardalis and the other 24 species in Syngnathidae using Solenostomus paradoxus as outgroup. Number above each node indicates the ML bootstrap support the probability generated from 100 replicates. The GenBank accession number for the complete mitochondrial genome was given following the species name.
Disclosure statement
The authors declare no conflict of interest in the preparation and execution of this manuscript.
Additional information
Funding
References
- Chen M, Zhu L, Chen J, Zhang G, Cheng R, Ge Y. 2017. The complete mitochondrial genome of the short snouted seahorse Hippocampus hippocampus Linnaeus 1758 (Syngnathiformes: Syngnathidae) and its phylogenetic implications. Conserv Genet Resour. 10:783–787.
- Cheng R, Fang Y, Ge Y, Liu Q, Zhang G. 2017a. Complete mitochondrial genome sequence of the Jayakar’s seahorse Hippocampus Jayakari Boulenger, 1900 (Gasterosteiformes: Syngnathidae). Mitochondrial DNA B. 2:593–594.
- Cheng R, Liao G, Ge Y, Yang B, Zhang G. 2017b. Complete mitochondrial genome of the great seahorse Hippocampus kelloggi Jordan & Snyder, 1901(Gasterosteiformes: Syngnathidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28:227–228.
- Cheng R, Zheng X, Ma Y, Li Q. 2013. The complete mitochondrial genomes of two octopods Cistopus Chinensis and Cistopus taiwanicus: revealing the phylogenetic position of the genus Cistopus within the order Octopoda. PLoS One. 8:e84216.
- Ge Y, Zhu L, Chen M, Zhang G, Huang Z, Cheng R. 2018. Complete mitochondrial genome sequence for the endangered Knysna seahorse Hippocampus capensis Boulenger 1900. Conserv Genet Resour. 10: 461–465.
- Hou F, Wen L, Cheng P, Guo J. 2018. Identification of marine traditional Chinese medicine dried seahorses in the traditional Chinese medicine market using DNA barcoding. Mitochondrial DNA A DNA Mapp Seq Anal. 29:107–112.
- Kumaravel K, Ravichandran S, Balasubramanian T, Sonneschein L. 2012. Seahorses - a source of traditional medicine. Nat Prod Res. 26:2330–2334.
- Lourie SA, Foster SJ, Cooper EWT, Vincent ACJ. 2004. A guide to the identification of seahorses. Project seahorse and TRAFFIC North America. Washington (DC): University of British Columbia and World Wildlife Fund.
- Wang Z, Ge Y, Cheng R, Huang Z, Chen Z, Zhang G. 2017. Sequencing and analysis of the complete mitochondrial genome of Hippocampus spinosissimus Weber, 1913 (Gasterosteiformes: Syngnathidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28:303–304.
- Zhu L, Chen M, Cheng R, Ge W, Zhang G, Ge Y. 2018. Complete mitochondrial genome characterization of the alligator pipefish Syngnathoides biaculeatus and phylogenetic analysis of family Syngnathidae. Conserv Genet Resour.DOI: 10.1007/s12686-018-1032-1.
