Abstract
Here, we present the complete mitochondrial genome of the Blotched snake, Elaphe sauromates (Pallas, 1814). Mitogenome complete sequence is 17,187 bp long and consists of 13 protein-coding genes (PCGs), two rRNA genes, and 22 tRNA genes and two control regions. The species mitochondrial genome has the same gene order as other mitogenomes of Elaphe spp. Their analysed genome has base composition as: 34.8% (A), 26.8% (C), 25.7% (T), and 12.7% (G), with an A + T bias (60.5%). Presented mitogenome will provide new data for phylogenetic analysis within the genus Elaphe.
Three species of the Eurasian genus Elaphe affect Western Palearctic. One of them is the Blotched snake, Elaphe sauromates (Pallas, 1814) considered for long time as a subspecies of Elaphe quatuorlineata (Bonnaterre, 1790). Its species level was confirmed by Utiger et al. (Citation2002). This snake currently ranging from the eastern Balkans and Asia Minor peninsula to western border of Central Asian region, Armenian Highland and the Levant (Sindaco et al. Citation2013).
Tissue sample (muscle from adult, no. 1179 stored in the collection of tissue samples at the Department of Zoology, Comenius University in Bratislava, Slovakia) was collected near Solenoe Ozero railway station (45.937 °N, 34.457 °E; in the limits of the species type locality defined by Pallas Citation1831), Crimea. Total genomic DNA was isolated by commercial DNA extraction kit (Qiagen DNeasy® Blood and Tissue Kit, Venlo, Netherlands) according to the manual. DNA library preparation (0.5 ng total DNA) was carried out according to the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA) workflow. Sequencing was performed with MiSeq Sequencing kit version 3 (Illumina, San Diego, CA) using Illumina MiSeq platform (2 × 200 bp).
For data analysis with CLC Genomics Workbench 9.5.2 (https://www.qiagenbioinformatics.com) app.9.7 million of paired-end sequencing reads were first trimmed using quality filter 0.01. De novo assembly into 213.000 contigs using strict parameters (Length fraction 0.8, Similarity fraction 0.9) followed by mapping to the mitochondrial genome of the most similar species Elaphe bimaculata (KM065513) enabled the identification of mitochondrial DNA represented by four sequences (267–11.587 bp) that were assembled into consensus sequence. Finally, gaps were filled manually mapping the reads (Length fraction 0.3, Similarity fraction 0.8) to the obtained consensus sequence.
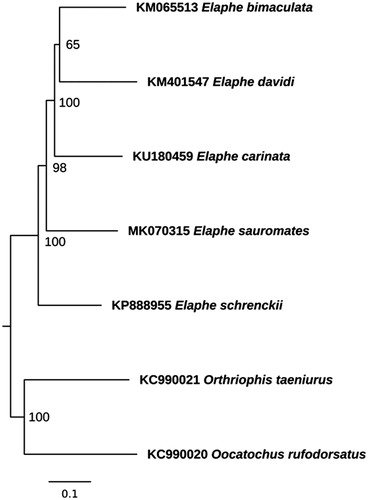
The phylogenetic analysis for whole mitogenome sequences of Elaphe was performed (). They were aligned using multiple sequence alignment program Muscle version 3.8.3.1 (Berkeley, CA, USA) (Edgar Citation2004). All gaps and poorly aligned positions were removed using Gblocks version 0.91b (Barcelona, Spain) (Talavera and Castresana Citation2007), resulting in 16,775 bp length alignment. The phylogenetic relationships were reconstructed using the maximum likelihood (ML) method in the PhyML version 2.4.5 (Montpellier, France) (Guindon and Gascuel Citation2003) and tested by 1000 bootstrap replications. The best substitution model was selected by the jModelTest version 2.1.10 (Vigo, Spain) (Darriba et al. Citation2012) based on the Bayesian information criterion (BIC).
Figure 1. Maximum likelihood phylogenetic tree of Elaphe representatives. The tree was created using TIM2 + I + G model. Mitogenomes of Orthriophis taeniurus (Cope, 1861) and Oocatochus rufodorsatus (Cantor, 1842) were used as outgroup. GenBank accession numbers and bootstrap values of nodes are shown on the tree.
The complete mitochondrial genome of E. sauromates has 17,187 bp (GenBank accession number: MK070315) and included 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and two control regions. The overall base composition of the genome in descending order was 34.8% – A, 26.8% – C, 25.7% – T, 12.7% – G, with an A + T bias (60.5%). Eleven of the 13 PCGs (ATP8, ATP6, COX2, COX3, CYTB, ND1, ND3, ND4, ND4L, ND5, ND6) used ATG as start codon, while COX1 used GTG and ND2 used ATT. Eleven genes (ATP8, ATP6, COX2, COX3, CYTB, ND1, ND2, ND3, ND4L, ND4, and ND5) ended with a TAA stop codon, but for six of them (COX2, COX3, CYTB, ND1, ND2, and ND3) TAA stop codon is completed by the addition of 3' A residues to the mRNA, ND6 gene ended with a AGG stop codon, and COX1 with AGA stop codon.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Pallas PS. 1831. Zoographia Rosso-asiatica. 3. Animalia monocardia seu frigidi sanguinis. Petropoli, Brazil: Caes. Academiae Scientiarum.
- Sindaco R, Venchi A, Grieco C. 2013. The reptiles of the Western Palearctic 2. Annotated checklist and distributional atlas of the snakes of Europe, North Africa, Middle East and Central Asia. Latina, Italy: Edizioni Belvedere.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577.
- Utiger U, Helfenberger N, Schätti B, Schmidt C, Ruf M, Ziswiler V. 2002. Molecular systematics and phylogeny of old and new world ratsnakes, Elaphe Auct., and related genera (Reptilia, Squamata, Colubridae). Russ J Herpetol. 9:105–124.
