Abstract
The complete chloroplast genome of Olmediella betschleriana was reconstructed by reference-based assembly using Illumina paired-end data. The assembled plastome is 158,171 base pairs (bp) in length, including a pair of inverted repeat regions (IRs) of 27,900 bp each, a large single-copy region (LSC) of 85,783 bp and a small single-copy region (SSC) of 16,588 bp. A total of 119 genes were predicted from the chloroplast genome, including 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content of O. betschleriana chloroplast genome was 36.71%. The phylogenetic analysis with 15 other species showed that O. betschleriana was closely clustered with Idesia polycarpa. The complete chloroplast genome of O. betschleriana provides new insight into Salicaceae evolutionary and genomic studies.
Olmediella betschleriana, which is native to Central America. It is an evergreen tree 25–40 feet tall with generally erect habit and low canopy that holds 4–6 inch long by 2.5 inch wide oval leathery dark green leaves that have spine-tipped teeth along the margins and are pinkish copper-coloured in new growth. Olmediella is classified in the heterogeneous family Flacourtiaceae (H.O. Sleumer Citation1980) but now it is placed Salicaceae (Chase et al. Citation2002) along with close relatives Bennettiodendron, Carrierea, Idesia, Itoa, Macrohasseltia, Poliothyrsis, and even the willows (Salix) and cottonwoods (Populus) themselves (Alford Citation2005). We characterized the complete chloroplast genome sequence of O. betschleriana to explore Salicaceae phylogenetic analysis. The annotated genome sequence has been deposited into Genbank with the accession number MK044831.
The voucher specimen of O. betschleriana was stored at Herbarium of Arnold Arboretum of Harvard University. We sampled an individual of O. betschleriana from Volcano of Santa Marta, Region of Los Tuxtlas, Veracruz and its altitude 1200 m. The total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle Citation1987). The whole-genome sequencing was generated with 350 bp pair-end reads on the Illumina Hiseq Platform. The programme NOVO-Plasty was used to assemble filtered reads (Dierckxsens et al. Citation2017) for O. betschleriana. The chloroplast sequence of Flacourtia indica (NC_037410.1) was used as a reference. Next, we annotated the assembled plastome by using Plann (Huang and Cronk Citation2015). Finally, we use Geneious (Kearse et al. Citation2012) to correct the annotation.
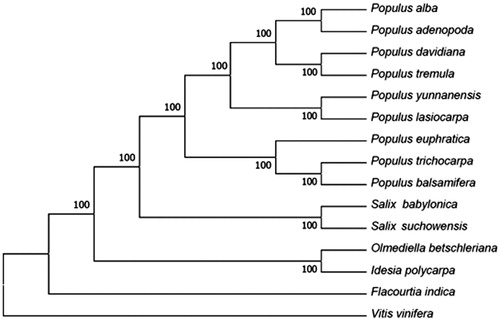
To dip into the phylogenetic position of O. betschleriana, 15 complete chloroplast genomes were downloaded from NCBI, including 13 popular and 2 other species. A Neighbour-Joining (NJ) tree was constructed using MEGA version 7.0 (Kumar et al. Citation2016). The phylogenetic tree showed that O. betschleriana and Idesia polycarpa were grouped together with a 100% bootstrap support ().
Figure 1. Phylogenetic tree of the complete chloroplast genome sequence of Olmediella betschleriana with other 15 species. The phylogenetic tree was constructed by MEGA version 7.0 software. The complete chloroplast genome sequences were downloaded from NCBI database. GenBank accession numbers: Idesia polycarpa (NC_032060.1), Populus davidiana (NC_032717.1), Populus rotundifolia (NC_033876.1), Populus tremula (NC_027425.1), Populus adenopoda (NC_032368.1), Populus alba (NC_008235.1), Populus yunnanensis (KP729176.1), Populus lasiocarpa (KX641589), Populus balsamifera (NC_024735.1), Populus trichocarpa (NC_009143.1), Populus euphratica (NC_024747.1), Salix suchowensis (NC_026462.1), Flacourtia indica (NC_037410.1), Salix babylonica (KT449800), and Vitis vinifera (NC_007957.1).
Our study provides further insights into Salicaceae evolutionary and genomic studies. The complete chloroplast genome of O. betschleriana was 158,171 bp in size and had 36.71% overall GC content. The chloroplast genome contained 119 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. And a pair of inverted repeat regions (IRs) of 27,900 bp each, a large single-copy region (LSC) of 85,783 bp and a small single-copy region (SSC) of 16,588 bp compose the plastome.
Disclosure statement
None of the co-authors has any conflict of interest to declare. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Alford MH. 2005. Systematic studies in Flacourtiaceae [dissertation]. Cornell (NY): University of New York.
- Chase MW, Zmarzty S, Lledo MD, Wurdack KJ, Swensen SM, Fay MF. 2002. When in doubt, put it in Flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull. 57:141–181.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: denovo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Sleumer. 1980. Flacourtiaceae. Flora Neotrop. 22:1–499. http://www.jstor.org/stable/4393727.
