Abstract
We have determined the mitochondrial genome of Dolichoderus sibiricus Emery 1889. The circular mitogenome of D. sibiricus was 16,086 bp including 13 protein-coding genes (PGCs), two ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 622 bp. The base composition was AT-biased (81.8%). Gene order of D. sibiricus is exactly same to Linepithema humile even though the two species belong to different tribes. Phylogenetic tree agrees with current phylogenetic placement of Dolichoderus in Dolichoderinae; while contradicts on relations of three subfamilies, Dolichoderinae, Myrmicinae, and Formicinae. D. sibiricus mitochondrial genome will be a good resource for further analyses.
The genus Dolichoderus is a diverse taxon with over 140 species occurring worldwide with the only exception of Saharan and sub-Saharan Africa (Shattuck and Marsden Citation2013). It is the sole member of tribe Dolichoderini and is the largest of all Dolichoderine genera (Bolton Citation2018). Dolichoderus sibiricus Emery 1889, which is a native species in East Asia, is a general scavenger species found foraging on tree trunks, forest floors, or sometimes tending aphids. Nests are found in dead branches or rotten wood are common in both forested and urban areas. They are easily distinguished by the four white spots on the first two gastral tergites, and the deep punctures covering from the head to the petiole.
As a first step to understand genomic characteristics of genus Dolichoderus, we completed mitochondrial genome of D. sibiricus isolated from Mt. Umyeonsan, Seoul, Republic of Korea. Genomic DNA was extracted from worker ants by using DNeasy Brood &Tissue kit (QIAGEN, Hilden, Germany) and was sequenced by Illumina HiSeq4000 (Macrogen, Inc., Seoul, Republic of Korea) . Raw sequences were filtered by Trimmomatic (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008). Gaps were filled by SOAPGapCloser 1.12 (BioMed Central, London, UK) (Zhao et al. Citation2011). AT-rich region was confirmed by Sanger sequencing of PCR product as well as all bases were confirmed by alignment against assembled genome (Li et al. Citation2009; Li Citation2013). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used to annotate its genome by comparing with that of Linepithema humile (KX146468). ARWEN (Laslett and Canbäck Citation2008) was used to annotate tRNAs. Ants were deposited in InfoBoss Cyber Herbarium (IN; Seoul, Republic of Korea; J. Park, KFDS00045).
Dolichoderus sibiricus mitochondrial genome length (Genbank accession is MH719017) is 16,086 bp. The nucleotide composition is AT-biased (81.8%). The mitogenome contains 13 protein-coding genes (PCGs), two rRNAs, and 22 tRNAs. The tRNAs size ranges from 63 to 75 bp, are similar to other ants (circa 54–90 bp). Despite that tribe of D. sibiricus (tribe Dolicoderini) is different from that of Linepithema humile (tribe Leptomyrmecini), gene order of two species is exactly same (Duan et al. Citation2016); while order of tRNA is different between D. sibiricus and Leptomyrmex pallens (Berman et al. Citation2014) and Dorymyrmex brunneus (MG253267) in tribe Leptomyrmecini. The control region presumably corresponds to the single largest non-coding AT-rich region (622 bp, A + T 94.4%).
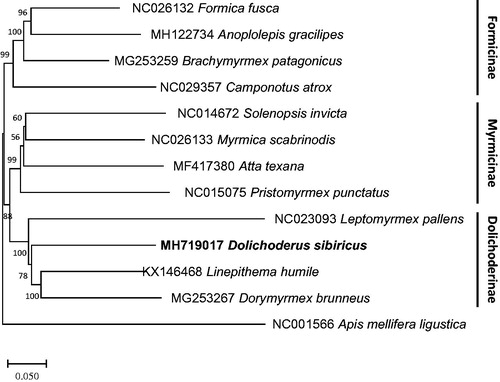
We inferred the phylogenetic relationship of 12 ant species including D. sibiricus, using concatenated alignments of nucleotide sequences of all PCGs. Multiple sequence alignments were conducted by MAFFT 7.388 (Katoh and Standley Citation2013) and were merged by Perl script. Neighbor joining tree was constructed using MEGA X with 10,000 bootstrap replicates (Kumar et al. Citation2018) presenting phylogenetic position of D. sibiricus based on mitochondrial genomes (). Leptomyrmex pallens (NC_023093) is positioned outside in subfamily Dolichoderinae clade, reflecting the gene order differences mentioned above (). Moreover, phylogenetic relation among three subfamilies, Dolichoderinae, Formicinae, and Myrmicinae, is different from the previous study (Ward Citation2014; ), indicating that more ant mitochondrial genome sequences are required to solve this relation correctly.
Figure 1. Phylogenetic tree of all available ants in Dorichoderinae subfamily: Dolichoderus sibiricus (This study; MH719017), Linepithema humile (KX146468), Leptomyrmex pallens (NC_023093), and Dorymyrmex brunneus (MG253267) as well as representative species of eight ant tribes:, Camponotus atrox (NC_029357), Anoplolepis gracilipes (MH122734), Formica fusca (NC_026132), Brachymyrmex patagonicus (MG253259), Myrmica scabrinodis (NC_026133), Solenopsis invicta (NC_014672), Atta texana (MF417380), Pristomyrmex punctatus (NC_015075), and Apis mellifera ligustica (NC_001566) as an outgroup. The numbers above branches indicate bootstrap support values.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Berman M, Austin CM, Miller AD. 2014. Characterisation of the complete mitochondrial genome and 13 microsatellite loci through next-generation sequencing for the New Caledonian spider-ant Leptomyrmex pallens. Mol Biol Rep. 41:1179–1187.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Bolton B. 2018. An online catalog of the ants of the world. [accessed 2018 May 03]. http://antcat.org.
- Duan XY, Peng XY, Qian ZQ. 2016. The complete mitochondrial genomes of two globally invasive ants, the Argentine ant Linepithema humile and the little fire ant Wasmannia auropunctata. Conserv Genet Resour. 8:275–277.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997.
- Shattuck SO, Marsden S. 2013. Australian species of the ant genus Dolichoderus (Hymenoptera: Formicidae). Zootaxa. 3716:101–143.
- Ward PS. 2014. The phylogeny and evolution of ants. Annu Rev Ecol Evol Syst. 45:23–43.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao QY, Wamg Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(14):S2.
