Abstract
Galliformes is an order of heavy-bodied ground-foraging birds, with important economic value and scientific value. The taxonomic statuses of Galliformes birds have been studied intensively; however, the phylogenetic relationships of some families within the Galliformes are still controversial. To demonstrate the phylogenetic relationships of the family in Galliformes, we analyzed the complete mitochondrial genome of 16 species belonging to seven families in Galliformes. The genetic distance among the seven families varied from 0.1466 (Megapodiidae and Tetraonidae) to 0.2594 (Megapodiidae and Odontophoridae). Maximum likelihood method was used to construct molecular phylogenetic tree. The phylogenetic tree grouped all the families into two deeply divergent clades. A monophyletic Odontophoridae was the sister-group to all other families in Galliformes. Our data well supported the megapodes and cracids were sibling taxa. Calibrated rates of molecular evolution, the divergent time of the seven families were 16–28 Myr.
Galliformes is an order of heavy-bodied ground-foraging birds. This order contains seven families: Megapodiidae, Cracidae, Odontophoridae, Numididae, Phasianidae, Meleagrididae, and Tetraonidae. They are important as seed dispersers and predators in the ecosystems they inhabit. The earliest galliform-like fossils hail from the Late Cretaceous, dating to about 85 million years ago (Myr). The taxonomic statuses of Galliformes birds have been studied intensively; however, the phylogenetic relationships of inter-familial within the Galliformes are still controversial. In the present study, we retrieved the complete mitochondrial genome sequences of seven families in Galliformes from GenBank. We used the molecular data to examine the interfamilial relationships in Galliformes.
A total of 16 species belonging to seven families in Galliformes were analyzed. The length of the complete mitochondrion genome ranged from 16,658 bp (Bonasa sewerzowi) to 16,748 bp (Acryllium vulturinum), with an average size of 16,694 bp. The nucleotide compositions were 24.52%T, 31.53%C, 30.27%A and 13.67% G, with a bias against G. The amount of A + T was more than that of G + C in complete mitochondrion genome among all the species. The genetic distance among the seven families varied from 0.1466 (Megapodiidae and Tetraonidae) to 0.2594 (Megapodiidae and Odontophoridae) (). Calibratedrates of molecular evolution, the divergent time of the seven families were 16–28 Myr.
Table 1. Genetic distance between families in Galliformes.
On the basis of hierarchical likelihood-ratio tests (hLRTs) as implemented in Modeltest 3.0 (Posada and Crandall Citation1998), GTR + G + I was used (-lnL = 80390.05, AIC = 160,858.11, BIC = 161,258.87). We set the proportion of invariant sites and the shape of the Gamma distribution as 0.34 and 0.79 (estimated by Modeltest), respectively. Maximum likelihood method was used to construct a molecular phylogenetic tree based on GTR + G + I model (). The phylogenetic tree grouped all the families into two deeply divergent clades (). Monophyly of the Odontophoridae clade was well supported by bootstrap analysis (BP = 100%), while the other clade contained the other families. All major clusters were supported by bootstrap values of 100% (). Mitochondrion genome data indicated Cracidae + Megapodiidae was sister taxa to Numididae (). The phylogenetic tree grouped the other three families (Meleagrididae, Phasianidae + Tetraonidae) into a subclade (, B2). Our data suggested the New World quail were not closely related to the Old World quail. Some molecular analyses had also supported this phylogeny, including DNA-DNA hybridization (Sibley and Ahlquist Citation1990). The New World quail (Odontophoridae) are morphologically and behaviorally distinct from the Phasianidae in many respects (Johnsgard Citation1999) and, thus, form a unique group within the Galliformes (Cox et al. Citation2007).
Figure 1. Phylogenetic tree of Galliformes constructed from complete mitochondrion genome. Numbers (in internodes) represent bootstrap values (=100%) from 1000 replications.
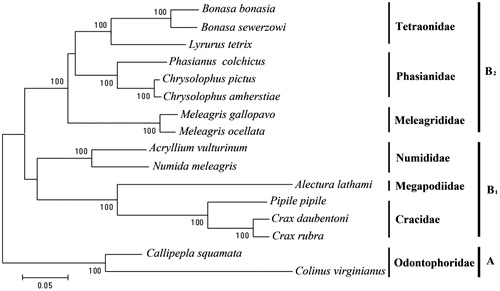
The evolutionary relationship between Megapodiidae has been an area of debate (Olson Citation1980). Olson (Citation1980) suggested that the megapodes might be cladistically relatively terminal, closer to the Phasianidae, but provided no cladistic evidence for this hypothesis. Sibley and Ahlquist (Citation1990) concluded that megapodes and cracids are sisters to one another. Mitochondrial genome well supported that the megapodes and cracids were sibling taxa (, BP = 100%).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cox WA, Kimball RT, Braun EL. 2007. Phylogenetic position of the New World quails: eight nuclear loci and three mitochondrial regions contradict morphology and the Sibley/Ahlquist tapestry. Auk. 124:71–84.
- Johnsgard PA. 1999. The Pheasants of the World, 2nd ed. Oxford: Oxford University Press.
- Olson SL. 1980. The significance of the distribution of the Megapodiidae. Emu. 80:21–24.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Sibley CG, Ahlquist JE. 1990. Phylogeny and Classification of Birds. New Haven: Yale University Press.
