Abstract
This study sequenced the complete mitochondrial genome of the female Amur leopard (Panthera pardus orientalis) using highest-quality (310× average coverage depth) big cat reference genomes. The total genome length was 16,964 base pairs, consisting of 13 protein-coding genes, 2 rRNAs, 22 tRNAs, and 1 control region. Phylogenetic analysis using protein-coding regions revealed that a P.p.orientalis was closely related to the P. pardus clade.
The leopard (Panthera pardus) is the most widespread big cat and among the five species in Panthera (Uphyrkina et al. Citation2001; Kitchener et al. Citation2017). Amur leopard (P.p.orientalis Schlegel, 1857; syn. P.p. japonensis Gray, 1862) is native to the Russian Far East and northern China, but extinct in the South Korea. Their decreasing range has led to a classification of “Critically Endangered” on the IUCN Red List (Stein et al. Citation2016; Kitchener et al. Citation2017). In this study, we sequenced the complete P.p.orientalis mitochondrial genome using Illumina HiSeq platforms. We then confirmed phylogenetic relationships among P.pardus using publicly available data from GenBank.
Tissue samples (cgrb15208) were collected from a dead female in the zoo at Daejeon O-World amusement park, South Korea and deposited in the Conservation Genome Resource Bank for Korean Wildlife(CGRB: www.cgrb.org), Seoul National University, Seoul, South Korea. A reference genome had been constructed previously, using the same samples (Kim et al. Citation2016). Protein-coding regions and rRNA were annotated with the online tool DOGMA (Wyman et al. Citation2004), while tRNA detection was performed in ARWEN (Laslett and Canback 2008)
The P.p.orientalis mitochondrial genome was 16,964 base pairs in size (GenBank MK043027), comprising 13 protein-coding genes, 22 tRNA, 2 rRNA, and 1 non-coding D-loop region. The light strand encoded the ND6 gene and nine tRNA genes while the heavy strand encoded all remaining genes. These mitogenome characters were similar to other Panthera species (Wei et al. Citation2011; Dou et al. Citation2016) and typical of vertebrate mitochondria (Saitoh et al. Citation2006).
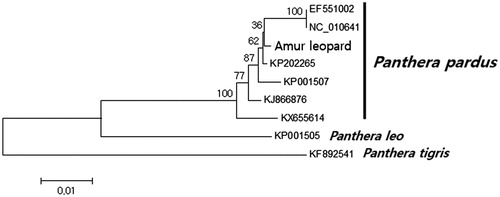
A neighbor-joining tree was constructed with 1000 bootstrap replicates in MEGA6.0 (Arizona State University, USA), using the 13 protein-coding genes of P.p.orientalis and 9 other Panthera species (). Our sequence was phylogenetically clustered with P. pardus. In conclusion, this study provides an important, high-quality data set for further molecular research on Panthera.
Figure 1. The consensus phylogenetic tree constructed using mitochondrial sequence data from a female Amur leopard and published Panthera data from GenBank. Numbers at the nodes are Bayesian posterior probability and neighbor-joining bootstrap values, estimated for concatenated sequences of 13 protein-coding genes and two ribosomal RNA genes.
Disclosure statement
The authors report no conflicts of interest and are alone responsible for the content and writing of the paper.
Additional information
Funding
References
- Dou H, Feng L, Xiao W, Wang T. 2016. The complete mitochondrial genome of the North Chinese Leopard (Panthera pardus japonensis). M DNA Part A. 27:1167–1168.
- Kim S, Cho YS, Kim HM, Chung O, Kim H, Jho S, Seomun H, Kim J, Bang WY, Kim C, et al. 2016. Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome Biol. 17:211.
- Kitchener AC, Breitenmoser-Wursten C, Eizirik E, Gentry A, Werdelin L, Wilting A, Yamaguchi N, Abramov AV, Christiansen P, Driscoll C, et al. 2017. A revised taxonomy of the felidae: the final report of the Cat Classification Task Force of the IUCN Cat Specialist Group. Cat News. Special Issue 11:1–80.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Saitoh l, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. 2006. Mitogenomic evolution and interrelationships of the cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63:826–841.
- Stein AB, Athreya V, Gerngross P, Balme G, Henschel P, Karanth U, Miquelle D, Rostro S, Kamler JF, Laguardia A. 2016. Panthera pardus. The IUCN Red List of Threatened Species 2016:e.T15954A50659089. Gland, Switzerland: IUCN.
- Uphyrkina O, Johnson WE, Quigley H, Miquelle D, Marker L, Bush M, O'Brien SJ. 2001. Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol Ecol. 10:2617–2633.
- Wei L, Wu X, Zhu L, Jiang Z. 2011. Mitogenomic analysis of the genus Panthera. Sci China Life Sci. 54:917–930.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
