Abstract
Zeugodacus tau (Walker) (Diptera: Tephritidae), is widely distributed throughout South and Southeast Asia. complete sequence of the mitogenome of Z. tau has been determined in this study. The circular genome is 15830 bp long and contains a standard gene complement, that is, the large and small ribosomal RNA subunits, 22 transfer RNA genes, 13 genes encoding mitochondrial proteins, and a non-coding A + T-rich control region. The phylogeny showed that Z. tau is closely related to Z. depressus with high bootstrap value supported and that monophyletic Bactrocera, Zeugodacus, and Ceratitidini were correctly identified within subfamily Dacinae.
Pumpkin fruit fly, Zeugodacus tau (Walker) (Diptera: Tephritidae), is widely distributed throughout South and Southeast Asia (White and Elson-Harris Citation1994). It is a polyphagous fruit pest and has been recorded on hosts from nine families, including Anacardiaceae, Cucurbitaceae, Fabaceae, Loganiaceae, Moraceae, Myrtaceae, Oleaceae, Sapotaceae, and Vitaceae (Allwood Citation1999). The species was previously placed under the subgenus Bactrocera (Zeugodacus), but was recently raised to generic rank (Virgilio et al. Citation2015). In this study, we reported the complete mitogenome of Z. tau from Kunming, Southwest China. The result would greatly facilitate phylogenetics, population genetics, and species identification.
The adult male flies were trapped from Kunming (25.05°N, 102.76°E), Southwest China, on 25 May 2018. Specimens were deposited in the museum of Southwest Forestry University (Voucher KM20180525), Kunming, China. Genomic DNA was extracted from adult fly following the manufacturer’s instruction in the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) and then sequenced and assembled using Illumina’s HiSeq2000 platform (Illumina, San Diego, CA). The sequence was preliminarily aligned within the CLUSTAL X program in BioEdit software. Protein-coding genes (PCGs), rRNA genes were predicted by using MITOS tools (Bernt et al. Citation2013), and tRNA were done through tRNAscan-SE (Lowe and Chan Citation2016).
The complete mitogenome of Z. tau is 15,830 bp long in size (GenBank MH900081). The base composition is 38.92% for A, 16.18% for C, 10.32% for G, and 34.58% for T. The mitogenome contains 13 PCGs, 22 transfer RNA genes, 2 ribosomal RNA genes, and a major non-coding region known as the CR (control region). J-strand codes nine PCGs (NAD2-3, NAD6, COX1-3, CYTB, ATP6, and ATP8) and 14 tRNAs, while N-strand codes four PCGs (NAD1, NAD4, NAD4l, and NAD5), eight tRNAs, and two rRNAs (16S rRNA and 12S rRNA). The gene arrangement of mitogenome is identical to the most common type of the putative ancestor of insects (Boore Citation1999; Cameron Citation2014).
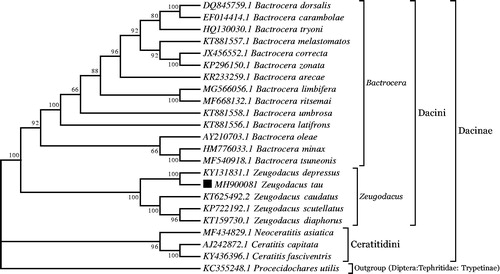
To validate the phylogenetic position, the mitogenomes of 25 species in subfamily Dacinae and the outgroup Procecidochares utilis (Diptera: Tephritidae: Trypetinae) were clustered together to construct maximum likelihood (ML) tree by using the ML method based on the Tamura-Nei model in MEGA 7 software (Kumar et al. Citation2016). The tree inferred from 500 replicates was taken to represent the phylogeny of the species analyzed in this study. The result indicated that Z. tau was closely related to Z. depressus with high bootstrap value supported (). Furthermore, three clades were correctly identified as assigned and monophyletic Bactrocera, Zeugoacus and Ceratitidini with high bootstrap confidence (). In conclusion, the mitochondrial genome of Z. tau educed in the present study can provide essential DNA molecular data for further phylogenetic and evolutionary analysis.
Figure 1. Molecular phylogeny for Zeugodacus tau and the related species in subfamily Dacinae based on complete mitogenome. Tree was constructed by maximum likelihood method with 500 bootstrap replicates. Genbank accession numbers lie before the scientific name of species. The position of Z tau is marked with solid square shape.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Allwood A. 1999. Host plant records for fruit flies (Diptera:Tephritidae) in South East Asia. Raffles Bull of Zool. 47:1–92.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation . Mol Phylogenet Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucl Acids Res. 44:54–57.
- Virgilio M, Jordaens K, Verwimp C, White IM, De Meyer M. 2015. Higher phylogeny of frugivorous flies (Diptera, Tephritidae, Dacini): localised partition conflicts and a novel generic classification. Mol Phylogenet Evol. 85:171–179.
- White IM, Elson-Harris MM. 1994. Fruit flies of economic significance: their identification and bionomics. Wallingford: CABI. Reprint with addendum.
