Abstract
Castanea henryi is Pearl chestnut, or the willow-leaved chestnut, which is widely distributed in the warm temperate subtropical climates of China. We determined the complete chloroplast genome sequence for C. henryi using Illumina sequencing data. The complete chloroplast sequence is 160,907 bp in length and contains a pair of 25,689 bp inverted repeats (IRs), a large single-copy (LSC) region of 90,531 bp, and a small single-copy (SSC) region of 18,998 bp. It contains 132 genes, including 102 protein-coding genes, eight ribosomal RNA genes (four RNA species), 46 transfer RNA genes, and two pseudogenes (ycf1 and ndhK). The overall GC content of the whole genome is 36.7%. Phylogenetic analysis based on 21 chloroplast genomes indicates that C. henryi is closely related to Chinese chestnut (C. mollissima) in Fagaceae.
Castanea henryi is belonging to the family Fagaceae and an endemic chestnut species native to the warm temperate subtropical climates of China. It is called as the willow-leaved chestnut or Pearl chestnut, the species is growing along the Yangtze River Valley and in Southern regions. It is well-known dry fruit that is indigenous to eastern and south-western China (Wu et al. Citation2012). It is cultivated for timber in Fujian and Zhejiang provinces, and the tree has been planted more than ever before in these areas in recent years (Xu et al. Citation2004; Wu et al. Citation2012). Some studies were reported the plant chemistry, Chinese medicine value, and nut stored value of C. henryi (Xu et al. Citation2004; Xu Citation2005; Wu et al. Citation2012). Despite this species’ value, existing genomic resources for C. henryi and genetic studies have been limited for Pearl chestnut. In this study, we report the complete chloroplast genome (cp) of C. henryi based on Illumina pair-end sequencing data.
The plant material of C. henryi was sampled from Nanchang, Jiangxi province, China. The voucher specimen is kept at the Evolutionary Botany Laboratory, College of Life Sciences, Northwest University (Xi’an, Shaanxi, China). Genomic DNA was extracted from leaf tissue using the Plant Genomic DNA kit (Tiangen Biotech, Beijing, China). The whole-genome sequencing was conducted with 350 bp pair-end reads on the IlluminaHiseq 4000 platform (Illumina, San Diego, CA) by Novogene, Beijing, China.
The high-quality paired-end reads were assembled with the program MITObim version 1.7 (Oslo, Norway) (Hahn et al. Citation2013) using the C. mollissima chloroplast genome sequence as a reference (GenBank accession HQ336406) (Jansen et al. Citation2011). Annotation was performed using the online program Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004). We generated a physical map of the chloroplast genome using OGDRAW (Lohse et al. Citation2013). The cp genome sequence was submitted to GenBank (accession number MH998384).
The cp genome of C. henryi was 160,907 bp in length and contains a pair of inverted repeats (IRa and IRb) regions of 25,689 bp, the large single-copy (LSC) region and small single-copy (SSC) region of 90,531 and 18,998 bp, respectively. The genome contains 132 genes, including 102 are protein-coding genes, 46 are transfer RNA genes, and eight ribosomal RNA genes. Among these genes, 15 genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhA, nahB, trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) have one intron, and three genes (clpP, rps12, and ycf3) have two introns. Overall GC content was 36.7% and in LSC, SSC, and IR regions were 34.6, 30.8, and 42.8%, respectively.
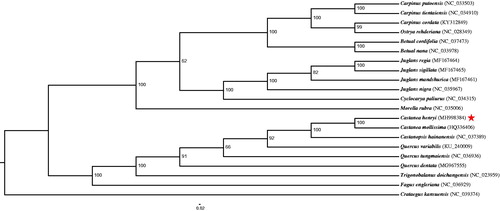
Plastome sequences of C. henryi plus seven Fagaceae, six Betulaceae, five Juglandaceae, and one Myricaceae species with one other plant species Crataegus kansuensis were aligned with MAFFT version 7.0.0 (Suita, Japan) (Katoh and Standley Citation2013). The phylogenetic relationship analysis was inferred using the model GTRGAMMA inference maximum likelihood (ML) method, which was performed using RAXML version 8.0 (Heidelberg, Germany) (Stamatakis Citation2014). The local bootstrap probability of each branch was calculated by 1000 replications. The phylogenetic tree showed that C. henryi was most closely related to Castanopsis hainanensis except C. mollissima with 100% bootstrap ().
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Jansen PK, Saski C, Lee SB, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28:835–847.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wu B, Zhang X, Wu X. 2012. New lignan glucosides with tyrosinase inhibitory activities from exocarp of Castanea henryi. Carbohyd Res. 355:45–49.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Xu J. 2005. The effect of low-temperature storage on the activity of polyphenol oxidase in Castanea henryi chestnuts. Postharvest Biol Technol. 38:91–98.
- Xu J, Zheng T, Meguro S, Kawachi S. 2004. Purification and characterization of polyphenol oxidase from Henry chestnuts (Castanea henryi). J Wood Sci. 50:260–265.