Abstract
Hibiscus syriacus L. belonging to family Malvaceae is a hardy deciduous shrub and is distributed in warm-temperature region. To understand the genetic diversity of ‘Mamonde’ cultivar, of which flower colour is white and its center is red, complete chloroplast genome sequence of H. syriacus L. ‘Mamonde’ was sequenced. Its length is 161,025 bp and has four sub-regions: 89,179 bp of large single copy (LSC) and 19,816 bp of small single copy (SSC) regions are separated by 25,745 bp of inverted repeat (IR) regions including 131 genes (86 coding regions, 8 rRNAs, and 37 tRNAs). The overall GC contents of the chloroplast genome were 36.8% and in the LSC, SSC, and IR regions were 34.7%, 31.1%, and 42.8%, respectively. This chloroplast genome will be a useful resource for understanding cultivar specific genetic variations of H. syriacus.
Hibiscus L. (Malvaceae) called rose of Sharon is a popular plant for ornamental plants, covering up to 350 species with many sections (Hochreutiner Citation1900; Bailey Citation1919; Bates Citation1965; Beers and Howie Citation1985; Pfeil and Crisp Citation2005). Genus Hibiscus is naive to warm temperate, subtropical, and tropical regions and its species diversity is concentrated in tropical areas (Pfeil et al. Citation2002). Hibiscus syriacus L., which is a fast-growing deciduous shrub, is a national flower of Republic of Korea. Its flower color is various and its blooming period is long enough even though that of individual flower is around one day (Kim and Fujieda Citation1991). There are seven major series of H. syriacus cultivars, presenting different development stages, flower shapes, and blooming period (Tolbert Citation1961; Kim and Fujieda Citation1991). ‘Mamonde’ cultivar has been established by breeding with Hongdansim and Baedal series. To understand the genetic background of this cultivar, we sequenced its whole chloroplast genome.
Fresh leaves of H. syriacus ‘Mamonde’ were used for extracting DNA using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Voucher specimen was deposited in InfoBoss Cyber Herbarium (IN); Y. Kim, IB-00569. Genome sequencing was performed using HiSeq4000 at Macrogen Inc. in Republic of Korea. We assembled chloroplast genome sequence with Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011) was used to fill gaps. We used Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) to annotate H. syriacus ‘Manonde’ chloroplast genome by transferring annotations from the congeneric H. syriacus sequence (NC_030195).
The chloroplast genome of H. syriacus ‘Mamonde’ (Genbank accession is MH330684) is 161,025 bp and has four sub-regions: 89,179 bp of large single copy (LSC) and 19,816 bp of small single copy (SSC) regions are separated by 25,745 bp of inverted repeat (IR). It contained 131 genes (86 coding regions, 8 rRNAs, and 37 tRNAs); 18 genes (seven coding regions, four rRNAs, and seven tRNAs) are duplicated in IR regions. The number of genes was increased in comparison to the previous report (Kwon et al. Citation2016) because annotation in IR regions was missed. The overall GC content of H. syriacus is 36.8% and in the LSC, SSC, and IR regions were 34.7%, 31.1%, and 42.8%, respectively.
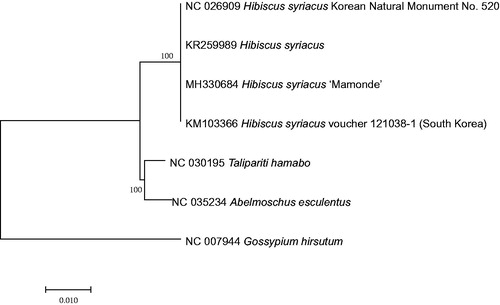
Because of reverse direction of SSC in both H. syriacus ‘Mamonde’ and Korean Natural Monument No. 520 (NC_026909; Kwon et al. Citation2016), occurred frequently in chloroplast genomes of Malvaceae (Chen et al. Citation2017), SSC sequences in both genomes were replaced and sequence alignment of seven chloroplast genomes covering H. syriacus and Talipaiti hamabo was conducted by MAFFT 7.388 (Katoh and Standley Citation2013). Neighbor joining phylogenetic tree was constructed using MEGA X with 10,000 bootstrap replicates (Kumar et al. Citation2018). The tree presents the phylogenetic position of H. syriacus among neighbor species (). This chloroplast genome sequences can be a resource for further understanding of genetic variations in various cultivars of H. syriacus.
Figure 1. Neighbor joining tree (bootstrap repeat is 10,000) of seven complete chloroplast genomes including four H. syriacus chloroplast genomes: H. syriacus (MH330684 (this study), NC_026909, KR259989, and KM103366), T. hamabo (NC_030195), Abelmoschus esculentus (NC_035234), and Gossypium hirsutum (NC_007944) as an outgroup. The numbers above branches indicate bootstrap support values of neighbor joining tree.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bailey LH. 1919. The standard cyclopedia of horticulture. Vol. 2. New York. USA: Macmillan.
- Bates DM. 1965. Notes on the cultivated Malvaceae: Hibiscus. Baileya. 13:56–130.
- Beers L, Howie R. 1985. Growing hibiscus. Kenthurst. Australia: Kangaroo Press.
- Chen Z, Grover CE, Li P, Wang Y, Nie H, Zhao Y, Wang M, Liu F, Zhou Z, Wang X, et al. 2017. Molecular evolution of the plastid genome during diversification of the cotton genus. Mol Phylogenet Evol. 112:268–276.
- Hochreutiner BPG, Hochreutiner BPG. 1900. Revision du genre Hibiscus. Genève. Swiss.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim J, Fujieda K. 1991. Studies on the flower color variation in Hibiscus syriacus L. J Korean Soc Hortic Sci. 32:247–255.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35:1547–1549.
- Kwon H-Y, Kim J-H, Kim S-H, Park J-M, Lee H. 2016. The complete chloroplast genome sequence of Hibiscus syriacus. Mitochondrial DNA Part A. 27:3668–3669.
- Pfeil B, Brubaker CL, Craven LA, Crisp M. 2002. Phylogeny of Hibiscus and the tribe Hibisceae (Malvaceae) using chloroplast DNA sequences of ndhF and the rpl16 intron. Syst Bot. 27(2):333–350.
- Pfeil B, Crisp M. 2005. What to do with Hibiscus? A proposed nomenclatural resolution for a large and well known genus of Malvaceae and comments on paraphyly. Australian Syst Bot. 18:49–60.
- Tolbert RJ. 1961. A seasonal study of the vegetative shoot apex and the pattern of pith development in Hibiscus syriacus. Am J Bot. 48:249–255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
