Abstract
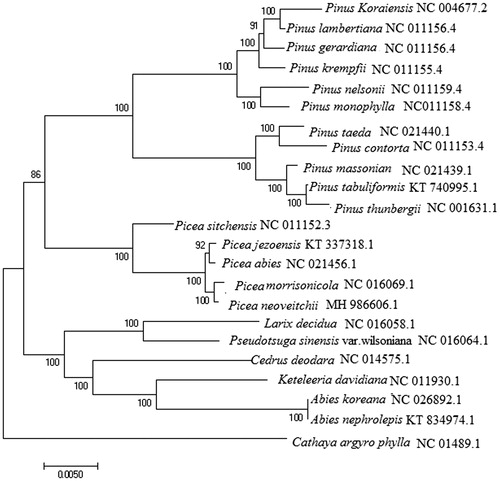
The complete chloroplast genome of Picea neoveitchii was assembled in this study. The genome comprised 124,234 bp in length. The GC content was 38.77%. A total of 116 genes are successfully annotated, including 77 protein-coding genes, 35 tRNA, and 4 rRNA genes. Eight protein-coding genes (rps12, trnV-UAC, rpoC1, atpF, rp12, trnI-GAU, trnL-UAA, and ycf3) contained one or two introns. Phylogenetic analysis showed that P. neoveitchii was most closely related to Picea morrisonicola.
Picea neoveitchii, a member of family Pinaceae, is an endemic species in China. It had been listed as an endangered species in red data books with scattering in several locations of south slope of the Qinling Mountain of China (Fu and Jin Citation1992; Zhang et al. Citation2006). Therefore, it is an urgent need for effective conservation of endangered species. Chloroplast has been a valuable tool to be used for phylogenetic studies due to its gene conservation and the lack of recombination (Ravi et al. Citation2008; Lin et al. Citation2012). P. neoveitchii is a timber tree of the Genus Picea which includes about 34 species, belongs to subfamily Piceoideae and found in the northern temperate and boreal regions (Farjon, Citation1990, Citation2010). To date, 22 complete chloroplast genomes have been reported for family Pinaceae. In this study, the complete chloroplast genome of P. neoveitchii (GenBank accession MH986606) was sequenced and analysed, aiming for further genetic diversity evaluation and phylogenetic analyses of the family Pinaceae.
The fresh, young leaves of P. neoveitchii was collected from Wudaoxia Nature Reserve, Hubei province China (110.08 E, 31.40 N). The specimen of P. neoveitchii was stored in the Xi’an Botanical Garden. Total genomic DNA was extracted with the DNeasy Tissue Kit (Tiangen, Beijing, China), and then used to construct a library for sequencing with Illumina Hiseq 2500 platform (Illumina, San Diego, CA). Additionally, MITObim version 1.8 (Hahn et al. Citation2013) was used to assemble the complete circular cp genome sequence. The cp genome was annotated and manually adjusted with CpGAVAS (Liu et al. Citation2012). The circular plastid genome map was completed with the help of the online programme OrganellarGenome DRAW (OGDRAW) (Lohse et al. Citation2013) and the annotated sequence was submitted to NCBI.
The complete cp genome sequence of P. neoveitchii was determined as 124,234 bp in length. The overall GC content was 38.77%. In total 116 genes were annotated, including 77 (66.38%) protein-coding genes (PCGs), 35 (30.17%) tRNA genes, and four (3.45%) rRNA genes. Seven protein-coding genes (rps12, trnV-UAC, rpoC1, atpF, rp12, trnI-GAU, and trnL-UAA) contained one intron, while ycf3 genes contained two introns.
Phylogenetic analysis was performed using the complete cp genomes of P. neoveitchii with those of 22 species in Pinaceae reported in Genbank of NCBI database by maximum likelihood method in MEGA version 7.0 (Kumar et al. Citation2016). The result demonstrated that P. neoveitchii was closely related to Picea morrisonicola and formed a sister group to the Picea clade ().
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Farjon A. 1990. Pinaceae: drawings and descriptions of the genera Abies, Cedrus, Pseudolarix, Keteleeria, Nothotsuga, Tsuga, Cathaya, Pseudotsuga, Larix and Picea. Konigstein, Germany: Koeltz Scientific Books.
- Farjon A. 2010. A hand book of the world’s conifers. Vol. 1–2. Leiden, Netherlands: EJ Brill.
- Fu LK, Jin JM. 1992. China plant red data book-rare and endangered plant. Beijing: Science Press; p. 538–539.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lin CP, Wu CS, Huang YY, Chaw SM. 2012. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 4:374–381.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW - a suite of tools for generating physicalmaps of plastid and mitochondrial genomes visualizing expression data sets. Nucleic Acids Res. 41:W575–W558.
- Ravi V, Khurana J, Tyagi A, Khurana P. 2008. An update on chloroplast genomes. Plant Syst Evol. 271:101–122.
- Zhang DS, Kim Y, Maunder M, Li XF. 2006. The conservation status and conservation strategy of Picea neoveitchii. Chin Popul Resour Environ. 4:58–64.