Abstract
Berchemiella wilsonii (Rhamnaceae) is an endangered endemic tree to China. Here, the chloroplast genome was characterized by next-generation Illumina paired-end sequencing. The complete genome comprised 160,076 bp in length, with an inverted repeat region of 26,514 bp, a small single copy (SSC) region of 18,717 bp, and a large single copy region of 88,331 bp. A total of 132 genes were identified, including 83 protein-coding genes, 37 transfer RNA genes, and eight rRNA genes. The overall GC content of the whole genome was 37.23%. Phylogenetic analysis showed that B. wilsonii was most closely related to Berchemiella berchemiifolia.
Berchemiella wilsonii, a member in the family Rhamnaceae, is one of the most critically endangered plants in China. The species has not been found for almost a hundred years since the first specimen collected originally by E. H. Wilson in the Xingshan county, Hubei Province, central China in 1907 and is treated as an extinct taxon, listed on the China Plant Red Data Book (Fu and Jin Citation1992). Due to anthropogenic activities such as habitat loss and deforestation, all taxa of the genus Berchemiella have been designated as nationally endangered species (Kang et al. Citation1991; Fu and Jin Citation1992; Iwatsuki et al. Citation1999). According to botanic expedition covering all recorded sites throughout its distribution range, five isolated adult trees of B. wilsonii were found in three sites in Hubei province(Kang et al. Citation2006). Therefore, it is an urgent need for effective conservation of endangered species, the critical importance for retaining the comprehensive genomics information of the species would definitely be used in attempting to formulate any conservation strategy. Thus, our study had got and the complete cp genome of B. wilsonii (GenBank accession number MH938366) to retrieve valuable resources for the conservation and phylogeny of B. wilsonii.
Four individual of B. wilsonii representing an isolated population was sampled from Wudaoxia Nature Reserve, Hubei, China (110.08 E, 31.40 N). The specimen of B. wilsonii was stored in the Harvard University Herbaria, its DNA was also stored in the Plant Gene Bank of Three Gorges Endangered Plant Species Resource Bank. Total genomic DNA was extracted from young and healthy leaves with the DNeasy Tissue Kit (Tiangen, Beijing, China), and then used to construct a library for sequencing with Illumina Hiseq 2500 platform (Illumina, San Diego, CA). The paired-end reads were trimmed for quality using the CLC Genomics Workbench version 9.0 (CLC Bio, Aarhus, Denmark) and NGSQCToolkit_version 2.3.3 with default parameters (Patel and Jain Citation2012). Additionally, MITObim version 1.8 (Hahn et al. Citation2013) was used to assemble the complete circular cp genome sequence. The cp genome was annotated and manually adjusted with CpGAVAS (Liu et al. Citation2012). The assembled genome was then manually checked for start and stop codons as well as intron/exon boundaries. The transfer RNA (tRNA) genes were further confirmed by tRNAscan-SE and ARAGORN (Lowe and Eddy Citation1997; Laslett and Canback, Citation2004; Schattner et al. Citation2005). Finally, the circular plastid genome map was completed using the online program OrganellarGenome DRAW (OGDRAW) (Lohse et al. Citation2013).
The complete cp genome of B. wilsonii was 160,076 bp in length presenting a typical quadripartite structure and contained an inverted repeat region (IR, 26,514 bp), a small single copy (SSC) region and a large single copy region (LSC) (18,717, 88,331 bp, respectively). Moreover, the cp genome encoded 132 genes, including 83 (62.88%) protein-coding genes (PCGs), 37 (28.03%) tRNA genes, and eight (6.06%) rRNA genes. Four genes (3.03%) were inferred to be pseudogenes. Fourteen genes (rpoC1, trnL-UAA, trnV-UAC, rpl22, rp12, ycf2, ycf15, trnI-GAU, ndhB, trnA-UGC, ndhA, trnA-UGC, trnI-GAU, and rps12) contained one intron, while three genes (ycf3, clpP, and ycf1) contained two introns. The overall GC content of the B. wilsonii cp genome was 37.23%, and the corresponding values in LSC, SSC, and IR regions were 35.09, 31.89, and 42.69%, respectively.
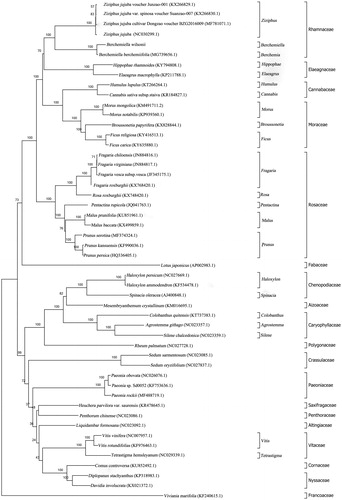
Phylogenetic analysis was performed using the chloroplast genome sequences of B. wilsonii and other 49 available cp genomes. A neighbour-joining phylogenetic tree was reconstructed with MEGA version 7 (Kumar et al. Citation2016) with 1000 bp replicate using the concatenated coding sequences of cp PCGs. The phylogenetic tree () showed that Rhamnaceae were monophyletic and sister to Elaegnaceae. Specifically, we observed that B. wilsonii was most closely related to Berchemiella berchemiifolia.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Fu LK, Jin JM. 1992. China plant red data book-rare and endangered plant. Beijing: Science Press; p. 538–539.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Iwatsuki K, Boufford DE, Ohba H. 1999. Flora of Japan. Vol. IIc. Tokyo: Kodansha Ltd.
- Kang SJ, Kim HE, Lee CS. 1991. Ecological studies on the distribution, structure and maintenance mechanism of Berchemia berchemiaefolia forest. Korean J Ecol. 14:25–38.
- Kang M, Zhang J, Wang J, Huang H. 2006. Isolation and characterization of microsatellite loci in the endangered tree Berchemiella wilsonii var. pubipetiolata and cross-species amplification in closely related taxa. Conserv Genet. 7:789–793.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32: 11–16.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW – a suite of tools for generating physicalmaps of plastid and mitochondrial genomes visualizing expression data sets. Nucleic Acids Res. 41:W575–W558.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PloS One. 7:e30619.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33:686–689.