Abstract
The comple mitogenome of Spanish flag snapper, Lutjanus carponotatus was determined by next generation sequencing (NGS) technique. The mitogenome of L. carponotatus (16,514 bp) habors 37 genes (22 tRNAs, 2 rRNAs, 13 polypeptides) and two non-coding regions, the origin of light strand replication (OL) and the putative control region (D-Loop). Among the 37 genes in L. carponotatus mitogenome, 28 were encoded on the H-strand, whereas only 9 were on the L-strand. Phylogenetic relationship showed that the full mitochondrial genome sequences L. carponotatus was most closely related to Lutjanus russellii with 93% sequence identity clustering with Lutjanus fulviflamma (92%) and Lutjanus vitta (91%).
The Spanish flag snapper, Lutjanus carponotatus (Richardson Citation1842), is distributed throughout the tropical and subtropical oceans worldwide, including Indo-west Pacific, India to northern Australia (Allen Citation1985; Newman et al. Citation2000). This species is important in recreational and commercial fisheries among the countries in the western tropical Pacific including Indonesia (Davis Citation1992; Kaunda-Arara and Ntiba Citation1997; Williams and Russ Citation1997). We, here, report the complete mitogenome sequence of L. carponotatus (GenBank Number: MK092066), which would help understand the biodiversity of fishes in genus Lutjanus.
The specimen was collected from the coastal water in Banyuwangi, Central Java, Indonesia (8°12'07,52"S 114°23'07,18"E), stored at Universitas Airlangga, Indonesia. Species identification of the specimen was confirmed by both its morphological charactericstics and DNA sequence identity in COI region (GenBank Number: KP194641). Mitochondrial DNA was extracted according to the manufacturer’s protocol by the mitochondrial DNA isolation kit ab65321 (Abcam, Cambridge, UK). Purified mitochondrial DNA was further fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA, USA). A library for sequencing was constructed by TruSeq® RNA library preparation kit V2 (Illumina Inc., San Diego, CA, USA) and its quality and the quantity was analyzed by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). DNA sequencing was performed by Illumina MiSeq sequencer (2 × 300 bp pair ends). The secondary structure of each tRNA was predicted by ARWEN program (Laslett and Canbäck Citation2008). The phylogenetic relationship of L. carponotatus complete mitogenome within family Lutjanidae was analyzed and constructed by software MEGA7 with minimum evolutionary (ME) algoritm (Kumar et al. Citation2016)
The complete mitochondrial genome of L. carponotatus was 16,514 bp in length, which contained 22 tRNAs, two ribosomal RNAs (12S and 16S), 13 polypeptides, and two non-coding regions; the origin of light strand replication (OL) and the putative control region (D-Loop). Among 37 genes, only 9 genes including (ND6 and 8 tRNA genes) were located on the L-strand, while the other 28 genes (two rRNAs, 12 protien coding genes, and 14 tRNAs) were encoded on the H-strand. Except for tRNASer, all the other tRNAs were able to form a three conserved tRNA cluster (IQM, WANCY, and HSL), which has typical stable stem-loop secondary structures (Satoh et al. Citation2016). ATG is used as start codon by all protein-encoding genes except for COX1 (GTG) and incomplete stop codons (TA-/T–) were identified in seven genes including ND2, ND3, ND4, ATP6, COX2,COX3, and Cytb. OL was located between tRNAAsn and tRNACys (WANCY region) and D-Loop was identified between tRNAPro and tRNAPhe.
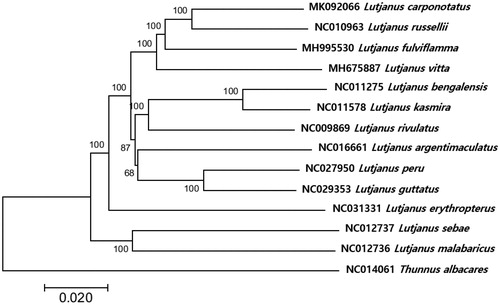
The phylogenetic tree with fish mitogenomes in genus Lutjanus showed that L. carponotatus was most closely related to Lutjanus russelli with 93% sequence identity followed by Lutjanus fulviflamma (92%) and Lutjanus vitta (91%) forming a clade (). This mitogenome information would help understand the genetic structures of the coral-reef dependent species for their effective conservation.
Figure 1. Phylogenetic relationship of Lutjanus carponotatus within family Lutjanidae. Phylogenetic analysis of Lutjanus carponotatus was constructed with the mitogenome sequences by MEGA7 software with Minimum Evolution (ME) algorithm with 1000 bootstrap replications. GenBank Accession numbers were shown followed by each scientific name. The mitogenome sequence of Thunnus albacares (NC014061) was used as an outgroup.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Allen GR. 1985. Review of the snappers of the genus Lutjanus (Pisces: Lutjanidae) from the Indo-Pacific, with the description of a new species. Indo-Pacific Fish. 11:1–87.
- Davis TL. 1992. Growth and mortality of Lutjanus vittus(Quoy and Gaimard) from the North West Shelf of Australia. Fish Bull. 90:395–404.
- Kaunda-Arara B, Ntiba M. 1997. The reproductive biology of Lutjanus fulviflamma (Forsskål, 1775) (Pisces: Lutjanidae) in Kenyan inshore marine waters. Hydrobiologia. 353:153–160.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Newman SJ, Cappo M, Williams DM. 2000. Age, growth and mortality of the stripey, Lutjanus carponotatus (Richardson) and the brown-stripe snapper, L. vitta (Quoy and Gaimard) from the central Great Barrier Reef, Australia. Fish Res. 48:263–275.
- Richardson, J. 1842. Contributions to the ichthyology of Australia. Annals and Magazine of Natural History ns 9:15–31.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17:719–738.
- Williams DD, Russ GG. 1997. Review of data on fishes of commercial and recreational fishing interest on the Great Barrier Reef. GBRMPAR Rep No. 33. I:1–113.
