Abstract
Stropharia rugosoannulata (Strophariaceae, Basidiomycota) is an edible mushroom distributed in northern temperate zones throughout the world. In this study, the complete mitochondrial genome sequence of S. rugosoannulata was determined. This mitochondrial genome is 60,551 bp in length with a GC content of 31.0% and encodes 14 conserved proteins, 2 ribosomal RNAs, 29 transfer RNAs, and 12 further ORFs. Phylogenetic relationships based on the mitochondrial genome between S. rugosoannulata and other basidiomycetes are inconsistent with those based on nuclear genes. This is the first report of the mitochondrial genome of mushrooms belonging to the family Strophariaceae.
Stropharia rugosoannulata is an edible mushroom belonging to the family Strophariaceae, which is distributed in northern temperate areas. Several studies on functional-food constituents of this mushroom, such as polysaccharides (He et al. Citation2012) and low-molecular weight compounds (Wu et al. Citation2011; Wu et al. Citation2012; Wu et al. Citation2013a; Wu et al. Citation2013b) have been reported. Recently, genome sequences of 90 mushrooms including S. rugosoannulata were determined and their phylogenetic relationships were analyzed based on all single-copy orthologous genes (Li et al. Citation2018). Although the mitochondrial genome has been widely used for fungal studies on molecular evolution and phylogenetics (Bullerwell and Lang Citation2005), little is known about the mitochondrial genome of the family Strophariaceae to which S. rugosoannulata belongs. In the present study, we determined the complete mitochondrial genome sequence of S. rugosoannulata and analyze its phylogenetic relationship with other basidiomycetes based on the mitochondrial genome.
The genomic DNA library of S. rugosoannulata strain NBRC 31871 (collected in Shiga, Japan) was constructed using the TruSeq DNA PCR-Free Sample Preparation Kit and sequenced for 301-bp paired-end reads using the MiSeq (Illumina, San Diego, CA). The raw sequence reads were cleaned up using Trimmomatic (Bolger et al. Citation2014) by trimming adapter sequences and low-quality ends (quality score, <10), and the reads derived from mitochondrial genome were extracted using khmer (Crusoe et al. Citation2015; Bankevich et al. Citation2012) with a high k-mer coverage (>100). The resultant 300,102 pair reads totaling 168.5 Mb were assembled using SPAdes ver. 3.12.0 (Bankevich et al. Citation2012). The complete mitochondrial genome of this mushroom is a circular DNA of 60,551 bp in length with a GC content of 31.0% (DDBJ/EMBL/GenBank: AP019006).
Gene prediction and annotation of the mitochondrial genome of S. rugosoannulata was performed using the MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and manually curated. Ribosomal RNA genes were predicted by aligning the mitochondrial genome to that of Coprinopsis cinerea (NW_003307477.1) using Mauve (Darling et al. Citation2004). The mitochondrial genome of S. rugosoannulata contained 57 genes including 26 protein-coding genes, two ribosomal RNA genes (rnl and rns), and 29 tRNA genes (for all 20 standard amino acids). The 26 protein-coding genes encoded 14 conserved mitochondrial proteins (cox1–3, cob, nad1–6, nad4L, atp6, atp8, and atp9), 9 endonucleases, a ribosomal protein S3, DNA polymerase, and hypothetical protein.
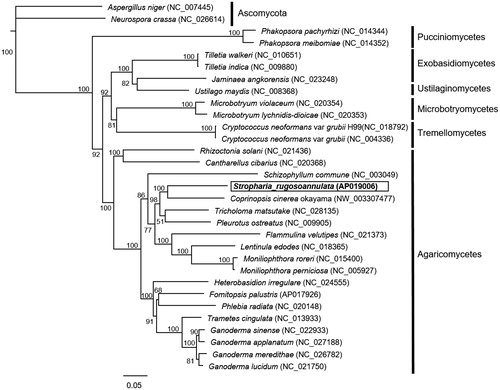
Phylogenetic analysis based on 9 proteins (cox1–3, cob, nad1–3, and nad5–6) conserved in 29 basidiomycetes and 2 ascomycetes was performed using Geneious version 9.1 (Kearse et al. Citation2012), as reported previously (Tanaka et al. Citation2017). The concatenated amino acid sequences were aligned using the MAFFT (Katoh et al. Citation2002) and the phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei Citation1987). Phylogenetic analysis based on the mitochondrial genome showed that S. rugosoannulata was closely related to C. cinerea (), which is inconsistent with nuclear gene-based phylogenetic relationships reported previously (Li et al. Citation2018). This is the first report on the mitochondrial genome of the family Strophariaceae, which will contribute to future studies on the evolution and classification of mushrooms.
Figure 1. Molecular phylogenetic analysis of 29 basidiomycetes and 2 ascomycetes. The phylogenetic tree was constructed by the neighbor-joining method based on concatenated amino acid sequences of 9 mitochondrial proteins (cox1–3, cob, nad1–3 and nad 5–6). Genbank accession numbers used in this analysis are provided next to each species name. Bootstrap values higher than 50 are shown at the nodes. Scale bar indicates the number of substitutions per site.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Bullerwell CE, Lang BF. 2005. Fungal evolution: the case of the vanishing mitochondrion. Curr Opin Microbiol. 8:362–369.
- Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S et al. 2015. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 4:900.
- Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403.
- He P, Geng L, Wang J, Xu CP. 2012. Production, purfication, molecular characterization and bioactivities of exopolysaccharides produced by the wine cap culinary-medicinal mushroom, Stropharia rugosoannulata 2# (higher Basidiomycetes). Int J Med Mushrooms. 14:365–376.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li H, Wu S, Ma X, Chen W, Zhang J, Duan S, Gao Y, Kui L, Huang W, Wu P, et al. 2018. The Genome Sequences of 90 Mushrooms. Sci Rep. 8:9982
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Tanaka Y, Suzuki T, Kurokura T, Iigo M, Toyama F, Habu N, Dohra H, Konno N. 2017. The complete genome sequence and phylogenetic analysis of the mitochondrial DNA of the wood-decaying fungus Fomitopsis palustris. Genes Genom. 39:1377–1385.
- Wu J, Fushimi K, Tokuyama S, Ohno M, Miwa T, Koyama T, Yazawa K, Nagai K, Matsumoto T, Hirai H, et al. 2011. Functional-food constituents in the fruiting bodies of Stropharia rugosoannulata. Biosci Biotechnol Biochem. 75:1631–1634.
- Wu J, Kobori H, Kawaide M, Suzuki T, Choi JH, Yasuda N, Noguchi K, Matsumoto T, Hirai H, Kawagishi H. 2013a. Isolation of bioactive steroids from the Stropharia rugosoannulata mushroom and absolute configuration of strophasterol B. Biosci Biotechnol Biochem. 77:1779–1781.
- Wu J, Suzuki T, Choi JH, Yasuda N, Noguchi K, Hirai H, Kawagishi H. 2013b. An unusual sterol from the mushroom Stropharia rugosoannulata. Tetrahedr Lett. 54:4900–4902.
- Wu J, Tokuyama S, Nagai K, Yasuda N, Noguchi K, Matsumoto T, Hirai H, Kawagishi H. 2012. Strophasterols A to D with an unprecedented steroid skeleton: from the mushroom Stropharia rugosoannulata. Angew Chem Int Ed. 51:10820–10822.
