Abstract
The complete mitochondrial genome of the Orbilia dorsalia was determined using Illumina sequencing. This mitogenome is a circular molecule of 191042 bp in length with a GC content of 25.3%, including 14 protein-coding genes, 26 transfer RNA genes, 2 ribosomal RNA genes. The phylogenetic analysis based on the sequences at the 14 concatenated mitochondrial protein-coding genes showed the relationships between O. dorsalia and other representative species is consistent with those based on nuclear genes.
Orbilia Fries, a globally distributed genus comprising more than 400 species, is connected to nematode-trapping fungi and known for its broad pleomorphism (Pfister Citation1997; Yu et al. Citation2011; Zhang and Hyde Citation2014). O. dorsalia was described as a new species (Yu et al. Citation2007) and the mature apothecia of the type strain can easily be formed where agar was removed and nematodes were added to induce trapping structures in artificial cultures. However, little is known about its mitogenome and its insights into sexual reproduction of this fungus. Here, we report the complete mitogenome of O. dorsalia and investigate the phylogenetic relationships with other related species based on mitochondrial proteins.
The mitogenome was extracted from the whole genome sequence of a pure culture of the type strain YMF1.01835 (collected in Yunnan province) using the Illumina HiSeq-1TB platform (Nanjing, China). This strain has been deposited in State Key Laboratory for Conservation and Utilization of Bio-Resources at Yunnan University. The mitogenome was identified by BLAST as scaffold 113, scaffold 130, and scaffold 137 in whole genome. The gaps were filled by separate PCR and sequencing with primers on regions flanking the gaps, resulting in one circular mitochondrial genome annotated using MFannot (http://megasun.bch.umontreal. ca/cgi-bin/mfannot/mfannot Interface.pl) and GLIMMER (https://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_ 3.cgi). The tRNAs were annotated using tRNAscan-SE (Schattner et al. Citation2005). All ORFs were searched and identified by ORFFinder (https://www.ncbi.nlm.nih.gov/orffinder/).
The complete mitogenome of O. dorsalia is a closed circular molecule of 191042 bp in length with a GC content of 25.3%, it consists of 14 core mitochondrial protein-coding genes, 26 tRNA genes, 2 rRNA genes (small and large subunit rRNA). Protein-encoding genes include three ATP synthase subunits (atp6, atp8, and atp9), three cytochrome oxidase subunits (cox1, cox2, and cox3), one apocytochrome b (cob), and seven NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). All the predicted genes totally possessed 40 introns, but none of the tRNAs possessed introns. All tRNA genes are encoded on the sense strand and covered 19 kinds of standard amino acids.
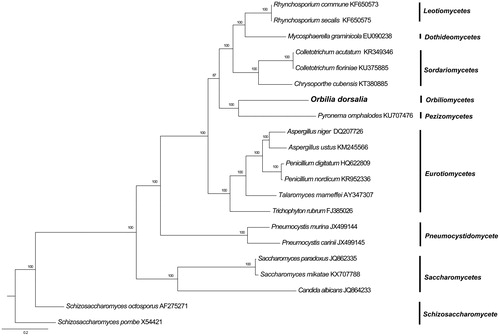
Phylogenetic analysis of O. dorsalia was performed by comparison with 14 mitochondrial proteins of other 21 species in Ascomycota was performed by Bayesian inference (BI). As shown in . Orbilia dorsalia was a member of Ascomycota and closely related to P. omphalodes, a result that is similar to previous researches (Jiang et al. Citation2018, Zhou et al. Citation2018). The phylogenetic relationship using mitochondrial proteins were in accordance with those constructed based on sequences of nuclear genes (Sugiyama et al. Citation2006). The Genbank Accession number for Orbilia dorsalia is MK547647.
Figure 1. Phylogenetic relationships among 21 Ascomycota fungi inferred based on the concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The following 14 mitochondrial protein-coding genes were concatenated: atp6, atp8, atp9, cytb, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6. The tree was generated using Bayesian inference (BI). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000,000 replicates) were shown next to the branches.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
References
- Jiang LL, Zhang YR, Xu J, Zhang KQ, Zhang Y. 2018. The complete mitochondrial genome of the nematode-trapping fungus Arthrobotrys oligospora. Mitochondrial DNA B. 3:968–969.
- Pfister D. 1997. Castor, Pollux and life histories of fungi. Mycologia. 89:1–23.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686
- Sugiyama J, Hosaka K, Suh S-O. 2006. Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia. 98:996–1005.
- Yu ZF, Qiao M, Zhang Y, Qin L, Zhang KQ. 2011. Pseudotripoconidium, a new anamorph genus connected to Orbilia. Mycologia. 103:164–173.
- Yu ZF, Zhang Y, Qiao M, Zhang KQ. 2007. Orbilia dorsalia sp. nov., the teleomorph of Dactylella dorsalia sp. nov. Crypt Mycol. 28:1–00.
- Zhang KQ, Hyde KD. 2014. Nematode-Trapping Fungi. Netherlands: Springer.
- Zhou DY, Zhang YR, Xu J, Jiang LL, Zhang KQ, Zhang Y. 2018. The complete mitochondrial genome of the nematode-trapping fungus Dactylellina haptotyla. Mitochondrial DNA B. 3:964–965.
