Abstract
In the present study, the complete mitochondrial DNA sequence of was determined for the first time. The mitochondrial genome of Lebiasina astrigata is a circular molecule of 16,899 bp in size and contains 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 2 control regions. The base composition is 30.02% for A, 15.34% for G, 27.24% for C, and 27.40% for T. All protein-coding genes started with Met, ND1,ND2, CO2, ATP8, ATP6, CO3, ND4L, ND5, and ND6 ended by TAA as a stop codon, CO1 ended by AGG as a stop codon, ND4 ended by AGA as a stop codon,CO3, Cytb ended by TAG. The lengths of 12S ribosomal RNA and 16S ribosomal RNA are 951 bp and 1695 bp, respectively. The length of control region is 1221 bp, ranging from 15,678 bp to 16899 bp. The complete mitochondrial genome sequence provided here would be useful for further understanding the evolution of ratite and conservation genetics of Lebiasina astrigata.
Lebiasina astrigata belongs to the family Lebiasina astrigata and the order Characiformes and mainly distributed in freshwater basins in northern Ecuador and southern Colombia. There were a few reports about its basic biology data including genetic information. In this study, we first determined the complete mitochondrial genome of Lebiasina astrigata, which would provide us the basic molecular data for further study on its systematics and conservation biology.
In the present study, one specimen Lebiasina astrigata chosen for mitochondrial genome analysis were collected from the freewater basin of Colombia (4°17′09″N, 72°47′29″W) and stored in a refrigerator of −80 °C in Zhejiang Engineering Research Center for Mariculture and Fishery Enhancement Museum. We got the total genomic DNA from three different individuals’ muscle tissue with the phenol-chloroform method (Barnett and Larson Citation2012). The calculation of base composition and phylogenetic construction was conducted by MEGA6.0 software (Tamura et al. Citation2013). The transfer RNA (tRNA) genes were generated using the program tRNAs-can-SE (Lowe and Eddy Citation1997). The complete mt genome of Lebiasina astrigata was 16,899 bp and has been deposited in GenBank with accession no. MH921292.
The mitochondrial genome of this species encoded 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs, and a non-coding control region, as those found in other Lebiasinida species (Boore Citation1999; Zhu et al. Citation2018a, Citation2018b). The nucleotide composition of the genome of Lebiasina astrigata is A 30.02, G15.34, C 27.24, and T 27.40%. Except for the ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) encoded on the light strand, all others genes were encoded on the heavy strand.
All genes use ATG as start codon, except COI use GTG, which is quite common in vertebrate mtDNA (Miya et al. Citation2001; Li et al. Citation2016). ND1, ND2, CO2, ATP8, ATP6, ND4L, ND5, and ND6 ended by TAA as a stop codon, COI ended by AGG as a stop codon, ND4 ended by AGA as a stop codon, ND3, Cytb ended by a single TAG. The 12S rRNA and 16S rRNA are 951 and 1695 bp, which are both located in the typical positions between tRNA-Phe and tRNA-Leu (UUR), separated by tRNA-Val.
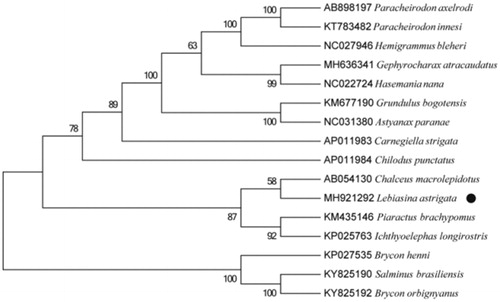
We performed bootstrap analyses (1000 replicates) to evaluate relative levels of support for various nodes in the phylogenie (). The NJ tree indicated two major groups of this genus and demonstrated that Lebiasina astrigata has a closest relationship with Chalceus macrolepidotus, which are consistent with the results based on morphology and other molecular methods (Inoue et al. Citation2010).
Figure 1. Neighbour joining (NJ) tree of 16 Lebiasinida species based on 12 PCGs. The bootstrap values are based on 10,000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Inoue JG, Miya M, Miller MJ, Sado T, Hanel R, Hatooka K, Aoyama J, Minegishi Y, Nishida M, Tsukamoto K. 2010. Deep-ocean origin of the freshwater eels. Biol Lett. 6:363–366.
- Kullander, SO, Fernholm, B. 1990. Check list of the freshwater fishes of south and central america. Copeia.2004(2004):714–716.
- Li Y, Xue D, Gao T, Liu J. 2016. Genetic diversity and population structure of the roughskin sculpin (Trachidermus fasciatus Heckel) inferred from microsatellite analyses: implicationsfor its conservation and management. Conserv Genet. 17:921–930.
- Lowe T, Eddy S. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhu K, Lu Z, Liu L, Gong L, Liu B. 2018a. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes:Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-j. 2018b. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA B. 3:536–537.
