Abstract
The complete mitochondrial genome of Therioaphis tenera (Aizenberg) was sequenced using Ion Torrent sequencing technology. The genome is 19,200 bp in length, which is the largest mitochondrial genome of aphids that has been sequenced so far. Therioaphis tenera mtDNA contains the D-loop, 36 coding genes instead of typical 37 due to the absence of Phe-tRNA, and a repeat region which is extremely long (3013 bp). Among all aphids with sequenced mitochondrial genomes, only T. tenera has a smaller number of genes in the mtDNA.
Therioaphis tenera (Aizenberg, 1956) is a specialist on different species of Caragana Fabr. and an invader in Belarus and other European countries (Zhorov et al. Citation2014). The territory of origin of T. tenera is Middle Asia where Caragana are native. Therioaphis tenera has been introduced to Europe together with its host-plants and is now widely distributed (Ripka Citation2004; Ratajczak et al. Citation2011; Sautkin and Buga Citation2012). Before the current work, no whole mitochondrial genomes of Calaphidinea have been published, which makes the T. tenera mtDNA especially interesting to study.
Imagines of T. tenera were collected 24.06.2015 in Belarus (N 53.932154; E 27.559755) from C. arborescens. Spacemen vouchers and morphological slides were included into the entomological collection of the Belarusian State University’s zoological museum.
The mitochondrial genome of T. tenera is 19,200 bp in length which is the largest known aphid mitogenome. It contains 13 protein-coding genes, two genes of ribosomal RNA and 21 tRNA genes. To date 18 aphid mitochondrial genomes were published, 15 of them concluded 37 genes, which is typical for animals, 1 did 38 and 1–39 genes due to the duplication of tRNAs (Ren et al. Citation2016). The Phe-tRNA is only a gene which is absent in the T. tenera mtDNA.
The T. tenera mitochondrial genome has a high A + T content of 79.1% that is slightly lower than in most of aphid mitogenomes, which usually is more than 80% (Wang et al. Citation2013). In protein-coding genes sequences, the A + T content is still 81.4%. Excluding the fact of one tRNA gene absence, the gene order is the same as in most of aphids, which is highly conservative. All protein-coding genes are initiated by the standard ATN codon and terminated with TAA, except the gene NAD4 with stop-codon TTT.
Aphid mitochondrial genomes have a regulatory region that is highly variable in length (Jager et al. Citation2014; Wang et al. Citation2014; Zhang et al. Citation2016) and usually is of 400–800 bp length but can vary from extremely short one in Hormaphis betulae (94 bp) (Li et al. Citation2017) to the largest in M. persicae (2531 bp). In the T. tenera mitochondrial genome, the regulatory region is 1451 bp long.
The repeat region was also found in T. tenera. Its repeat region is the largest of all known aphid repeat regions and contains two identical sequences of 161 bp each separated with a 132 bp region and with a 2553 bp non-coding region lying downstream. This type of repeat region is unusual in aphids because of both an extreme large size and a low proportion of A + T content (Ren and Wen Citation2017).
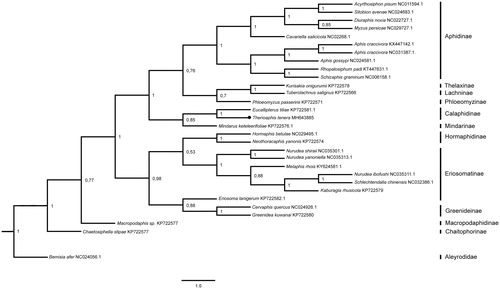
We used Bayesian analysis and MCMC method to estimate the phylogeny of aphids using the PCG of all 17 whole and 12 partial aphid mitogenomes available currently. In this phylogeny, all aphids were divided into two big groups (). Therioaphis tenera and another member of Calaphidinae together with Aphidinae, Thelaxinae, Lachninae, Ploeomyzinae, and Mindarinae separated from other aphids.
Nucleotide sequence accession number
The complete mitogenome sequence of T. tenera has been assigned GenBank accession number MH643885.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jager L, Burger NFV, Botha AM. 2014. Complete mitochondrial genome of Diuraphis noxia (Hemiptera: Aphididae) from nine populations, SNP variation between populations, and comparison with other Aphididae species. African Entomol. 22:847–862.
- Li YQ, Chen J, Qiao GX. 2017. Complete mitochondrial genome of the aphid Hormaphis betulae (Mordvilko) (Hemiptera: Aphididae: Hormaphidinae). Mitochondrial DNA Part A. 28:265–266.
- Ratajczak J, Wilkaniec B, Wilkaniec A. 2011. Infestation of dendrological collection in central Poland by aphids. Dendrobiology. 66:85–97.
- Ren ZM, Bai X, Harris AJ, Wen J. 2016. Complete mitochondrial genome of the Rhus gall aphid Schlechtendalia chinensis (Hemiptera: Aphididae: Eriosomatinae). Mitochondrial DNA Part B. 1:849–850.
- Ren ZM, Wen J. 2017. Complete mitochondrial genome of the North American Rhus gall aphid Melaphis rhois (Hemiptera: Aphididae: Eriosomatinae). Mitochondrial DNA Part B. 2:169–170.
- Ripka G. 2004. Recent data to the knowledge of the aphid fauna of Hungary (Homoptera: Aphidoidea). Acta Phytopathol Entomol Hungar. 39:91–97.
- Sautkin FV, Buga SV. 2012. List and the damaging potential of pests of Caragana (Сaragana arborescens lam.) in Belarus. Vestnik BSU. 2:90–91.
- Wang Y, Huang XL, Qiao GX. 2013. Comparative analysis of mitochondrial genomes of five Aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE. 8:e77511.
- Wang Y, Huang XL, Qiao GX. 2014. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae): the mitogenome of Cervaphis quercus. Insect Sci. 21:278–290.
- Zhang B, Zheng J, Liang L, Fuller S, Ma C-S. 2016. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae). Mitochondrial DNA. 27:945–946.
- Zhorov DG, Sautkin FV, Buga SV. 2014. Distribution of Therioaphis tenera (Aizenberg, 1956) (Sternorrhyncha: Drepanosiphidae) under the conditions of green stands In Belarus. Proc Belarusian State University. 9:124–129.