Abstract
We report the complete mitogenome of Sitona obsoletus, an agricultural pest in New Zealand and some European countries. Like other Sitona mitogenomes, the 6 tRNA gene box is ordered RNSAEF, supporting the hypothesis that this signature is common to, and potentially diagnostic, of this genus. The Trojan Female Technique (TFT) is a genetic pest control strategy that exploits mitochondrial DNA alleles that affect male, but not female fertility and fitness. The complete mitogenome is an essential first step in exploring the utility of TFT for the control of S. obsoletus.
The genus Sitona (Coleoptera: Curculionidae, Entiminae) contains over 100 species; all feed on legumes, and many are pests (Velazquez De Castro et al. Citation2007). Sitona obsoletus (Gmelin Citation1790), a Palearctic weevil, was accidentally introduced to several countries and is a pest of clovers in New Zealand (Basse et al. Citation2015). It is the focus of research to implement a species-specific method of genetic pest control – the Trojan Female Technique (TFT) (Wolff et al. Citation2017). TFT exploits naturally occurring mitogenome alleles that induce male specific sterility. To help identify such alleles in S. obsoletus, we sequenced and analyzed its mitochondrial genome.
Genomic DNA (gDNA) was extracted (Gemmell and Akiyama (Citation1996) (LiCl substituted with NaCl (6 M)) from a whole female (see Geolocation information). A PCR-free library (insert size of approximately 450 bp) was then constructed using a TruSeq DNA PCR-Free kit (Illumina). The library was run on a single rapid run lane of a Hiseq2500 (Illumina) with 250 bp paired-end reads. Output data was quality checked using FastQC (v0.11.5) and adaptor trimmed with cutadapt (v1.14). NOVOPlasty (v2.7.1; Dierckxsens et al. (Citation2016)) was used to assemble the mitogenome, with the Sitona callosus mitogenome (GenBank Accession number: MF594624.1) (Zhang et al. Citation2017) as the seed. Default settings were used except ‘Genome Range’ was set to ‘12000–32000’ (bp). We annotated the mitogenome using MITOS (v1) (Bernt et al. Citation2013) and manually edited/added stop codons/signals (GenBank Accession number: MH814932). The mitogenome is 17,003 bp long and, like other eukaryotes, includes one control region, two ribosomal RNA genes, 13 protein-coding genes (PCGs), and 22 tRNA genes. Gene content and order are identical to the canonical insect mitogenome (Cameron Citation2014), except that the six tRNA genes box is modified from ARNSEF to RNSAEF. This rearrangement may be a signature unique to Sitona weevils.
Beast2 (v2.4.8) (Bouckaert et al. Citation2014) was used to construct a phylogeny from published Entiminae mitogenomes, our S. obsoletus mitogenome, and that of an outgroup, Hypera postica (Hyperinae). We used a TN93 GAMMA (6 categories) site model, relaxed lognormal clock model, and a Yule tree model, for the 13 PCGs (concatenated and codon-partitioned). PCGs were aligned by translation using all available algorithms in Geneious (v11.1.5). The alignment with the highest average pairwise similarity (or identical sites % if the former values were equal) was selected for each gene. Two chains of 100,000,000 states, sampling every 10,000 were checked for convergence and stationarity using Tracer (v1.7.1) (Rambaut et al. Citation2018) and convergence using the R (v3.3.3) (R Core Team Citation2018) package RWTY (Warren et al. Citation2017) using RStudio (v 1.1.456) (RStudio Citation2015), and then visualized using FigTree (v1.4.3) (Rambaut Citation2017). The S. obsoletus mitogenome sits within a strongly-supported clade comprising all published Sitona mitogenomes ().
Figure 1. Phylogenetic tree of the Sitona obsoletus mitogenome with other published mitogenomes from the subfamily Entiminae and Hypera postica (Hyperinae) as an outgroup. Species’ mitogenome GenBank accession numbers are listed here according to their order in the figure from top to bottom: JN163953.1, MH814932, JN163948.1, MF594624.1, JN039360.1, MF807224.1, JN163949.2, JN163969.1, KX087368.1, JX412782.1, JX412810.1, KX087356.1, JN163950.1, MF383367.1 and NC_018354.1. Numbers near nodes indicates the posterior probability values for each and the scale bar shows the rate of substitution.
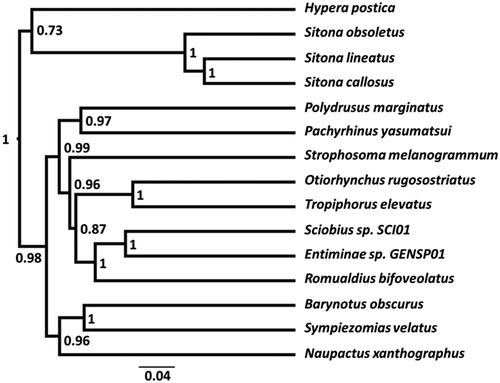
The S. obsoletus mitogenome provides further evidence that the Sitona six tRNA gene box signature could be a useful diagnostic tool for this genus, will aid future phylogenetic studies, and will be invaluable for the studies necessary to develop TFT for controlling S. obsoletus.
Geolocation information
The adult weevil was collected from a dairy farm on Rakaia Island (−43.845, 172.177), in the Rakaia River south of Southbridge, Canterbury, New Zealand.
Acknowledgements
We thank Dr Scott Hardwick (AgResearch, Lincoln) for collecting the sequenced weevil, Colin Ferguson (AgResearch, Invermay) for collecting other weevils for DNA extraction trials, and Dr Ela Sawicka (AgResearch, Lincoln) for helping to optimize the extraction protocol. We also thank Dr Hugh Cross (University of Otago, New Zealand) and Natalie Forsdick (University of Otago, New Zealand) for kindly recommending NOVOPlasty.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Basse B, Phillips C, Hardwick S, Kean J. 2015. Economic benefits of biological control of Sitona obsoletus (clover root weevil) in Southland pasture. NZ Plant Prot. 68:218–226.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mo Phylogen Evol. 69:313–319.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. System Entomol. 39:400–411.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18–e18.
- Gemmell NJ, Akiyama S. 1996. An efficient method for the extraction of DNA from vertebrate tissues. Trends Genet. 12:338–339.
- Gmelin J. 1790. Systema Naturae, ed. 13, I (4). Leipzig: Beer; [accessed 2018 Sep 1]. http://edoc.hu-berlin.de/docviews/abstract.php?lang=ger&id=26828
- Haran J, Timmermans MJ, Vogler AP. 2013. Mitogenome sequences stabilize the phylogenetics of weevils (Curculionoidea) and establish the monophyly of larval ectophagy. Mol Phylogen Evol. 67:156–166.
- R Core Team. 2018. R: A Language and Environment for Statistical Computing.
- Rambaut A. 2017. FigTree-version 1.4. 3, a graphical viewer of phylogenetic trees.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 10.
- RStudio T. 2015. RStudio: integrated development for R Boston, MA: RStudio, Inc.
- Velazquez De Castro AJ, Alonso ‐Zarazaga MÁ, Outerelo R. 2007. Systematics of Sitonini (Coleoptera: Curculionidae: Entiminae), with a hypothesis on the evolution of feeding habits. System Entomol. 32:312–331.
- Warren DL, Geneva AJ, Lanfear R. 2017. RWTY (R We There Yet): an R package for examining convergence of Bayesian phylogenetic analyses. Mol Biol Evol. 34:1016–1020.
- Wolff JN, Gemmell NJ, Tompkins DM, Dowling DK. 2017. Introduction of a male-harming mitochondrial haplotype via ‘Trojan Females’ achieves population suppression in fruit flies. eLife. 6:e23551.
- Zhang L, Wang J, Yang X-Z, Li X-P, Feng R-Q, Yuan M-L. 2017. Mitochondrial genome of Sitona callosus (Coleoptera: Curculionidae) and phylogenetic analysis within Entiminae. Mitochondrial DNA Part B. 2:538–539.
