Abstract
Medicago sativa cv. Hangmu No.1 is the first variety of alfalfa bred by space mutation breeding in China, which characterized by high multifoliate rate, high yield, and abundant nutrients. To some extent, this space mutation alfalfa will efficiently solve the severe shortage of forage and contribute to rapid development in animal husbandry in China. In this study, the complete chloroplast genome sequence of M. sativa cv. Hangmu No.1 was evaluated based on Illumina Hiseq 2500 platform. The circular genome was 123,825 bp in length, containing 102 genes including 75 protein-coding genes (70 PCG species), 24 transfer RNA genes (21 tRNA species) and 3 ribosomal RNA genes (3 rRNA species). Consistent with other Medicago chloroplast genomes, the M. sativa cv. Hangmu No.1 genome did not contain inverted repeat (IR) regions. Phylogenetic analysis based on 11 chloroplast genomics indicates that M. sativa cv. Hangmu No.1 is closely related to M. sativa and Medicago falcata strain. This complete chloroplast genome will provide valuable information for the molecular breeding of M. sativa.
Medicago sativa known as the king of forage crops has high production, strong resistance, and high nutritional value for livestock (Liu et al. Citation2013). It has been widely cultivated around the world and its planting time in China is over two thousand years (Bouton et al. Citation2007). Medicago sativa cv. Hangmu No.1 belongs to M. sativa and is cultivated by space breeding technology, which characterized by high hay yield, abundant crude protein, and amino acid. Most of this species has 5 leaves and its multifoliate rate is up to 41.5%, the crude protein and amino acid content reach 20.08% and 12.32%, respectively. Now M. sativa cv. Hangmu No.1 has been listed as a species and approved by the Gansu grass variety Committee (Accession: GCS014), China (Yang et al. Citation2015). However, genetic diversity of M. sativa cv. Hangmu No.1 has not yet been reported. In this study, we determined the complete chloroplast genome sequence of M. sativa cv. Hangmu No.1 based on Illumina Hiseq 2500 platform (San Diego, CA, USA) to provide a valuable complete chloroplast genomic information and illustrated the phylogeny of Hangmu No.1 based on the complete chloroplast genome sequences as well.
The fresh healthy leaves tissue was sampled from Lanzhou City, Gansu province, China (E103°49′54ʺ, N36°03′42ʺ). The voucher specimens of M. sativa cv. Hangmu No.1 were deposited in the Plant Germplasm Resources Bank of Shrub and Forage in Alpine and Arid Regions of Chinese. Total genomic DNA extraction and genome sequence assembling were conducted by Novogene Corporation (Beijing, China). The locations of protein-coding genes were determined by comparing with the corresponding sequences of other M. sativa species.
The complete assembled chloroplast genome of M. sativa cv. Hangmu No.1 was 123,825 bp in length, which encoded a total of 102 genes including 75 protein-coding genes (70 PCG species), 24 transfer RNA genes (21 tRNA species) and 3 ribosomal RNA genes (5S, 18S, and 28S rRNA). The majority of the above genes were single copy, but 3 PCG genes (ycf3, rpl14, and rpl2) and 1 tRNA gene (trnM-CAU) were duplicated. However, the chloroplast genome absents a typical quadripartite structure, which is consistent with other Medicago plants (Tao et al. Citation2017). The base composition of the genome is asymmetric (33% A, 16.43% G, 17.52 C and 33.06% T) with an overall GC content of 33.94%.
To reveal the phylogenetic position of M. sativa cv. Hangmu No.1, a neighbour-joining (NJ) phylogenetic tree was constructed with MEGA6 (Tamura et al. Citation2013) using the coding sequences of 11 species. We found that M. sativa cv. Hangmu No.1 was clustered with other Medicago with 100% bootstrap values and had a closer relationship to M. sativa and Medicago falcata strain ().
Figure 1. Phylogenetic relationships of 11 species based on complete chloroplast genome using the neighbour-joining methods. The bootstrap values were based on 1000 replicates and are shown next to the branches.
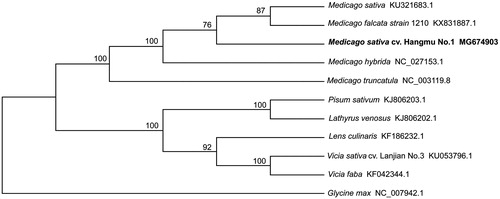
This study provided the characterization of the complete chloroplast genome structure of M. sativa cv. Hangmu No.1 and showed the phylogenetic relationships in Legume. All information about M. sativa cv. Hangmu No.1 will provide a fundamental genetic reference for future breeding programs of M. sativa.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bouton J, Conner AJ, Mercer CF. 2007. The economic benefits of forage improvement in the United States. Euphytica. 154:263–270.
- Liu Z, Chen T, Ma L, Zhao Z, Zhao PX, Nan Z, Wang Y. 2013. Global Transcriptome sequencing using the illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. Plos One. 8:e83549.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tao X, Ma L, Zhang Z, Liu W, Liu Z. 2017. Characterization of the complete chloroplast genome of alfalfa (Medicago sativa) (Leguminosae). Gene Rep. 6:67–73.
- Yang HS, Chang GZ, Zhou XH. 2015. Performance of Hangmu No.1 alfalfa in the Lanzhou region. Acta Prataculturae Sinica. 24:1138–1145.
