Abstract
The complete mitochondrial of Black-chinned Yuhina is 17,817 bp in length and contained 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and 2 control regions. The overall base composition of the mitogenome was 30.11% A, 24.12% T, 14.37% G, 31.41% C. All the genes in the Black-chinned Yuhina mitogenome were encoded on the H-strand, except for the ND6 and eight tRNA genes that were transcribed from the L-strand.
Black-chinned Yuhina (Yuhina nigrimenta), belonging to the genus Yuhina of the Timaliidae, is mainly distributed in southeastern and western China, India, Bangladesh, Bhutan, Cambodia, Laos, Myanmar, Nepal, and Thailand (Moyle et al. Citation2012), and it is listed as a Least Concern species in the Red List by the International Union for the Conservation of Nature (BirdLife International Citation2016). There is a controversial issue about the taxonomy of Black-chinned Yuhina. Some researchers classified Black-chinned Yuhina into the family Timaliidae (Cheng Citation1987; Gelang et al. Citation2009), while the other researchers categorized Black-chinned Yuhina into the family Zosteropidae (Cibois et al. Citation2002; Gill and Donsker Citation2018). In this study, we sequenced the complete mitogenome of Black-chinned Yuhina and analyzed these data, and except that it could provide molecular data for the taxonomy of Black-chinned Yuhina.
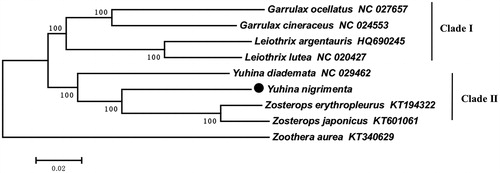
The sample was collected from Ya’an (30°0′47.20ʺN, 103°02′25.51ʺE), Sichuan province of China and stored at the Zoology laboratory, Sichuan Agricultural University, China. The total genomic DNA was extracted from the muscle tissue by using the standard phenol/chloroform methods (Sambrock and Russel Citation2001), and 15 pairs of primer were used to amplify the overlapping segment of its mitochondrial genome based on the Polymerase Chain Reaction (PCR). We provided the mitogenome sequence information of Black-chinned Yuhina and got the GenBank no. MH916608. For using the MEGA5.0 for neighbour-joining (NJ) analysis to construct a phylogenetic tree (Tamura et al. Citation2011), we downloaded eight mitochondrial genome sequences of other birds from the GenBank of Garrulax ocellatus (NC_027657), Garrulax cineraceus (NC_024553), Leiothrix argentauris (HQ690245), Leiothrix lutea (NC_020427), Yuhina diademata (KT340629), Zosterops erythropleurus (KT194322), Zosterops japonicas (KT601061), and Zoothera aurea (KT340629) was used as the outgroup.
The complete mitogenome sequence of Black-chinned Yuhina was 17,817 bp in length and contained 13 protein-coding genes (PCGs), 2 ribosomal RNA (12srRNA, 16srRNA) genes, 22 transfer RNA (tRNA) genes and 2 control regions (Dloop1, Dloop2) which was similar to other mitochondrial genomes of Passeriformes (Li et al. Citation2017; Zhou et al. Citation2016). The overall base composition of the mitogenome was 30.11% A, 24.12% T, 14.37% G, 31.41% C. All the genes in the Black-chinned Yuhina mitogenome were encoded on the H-strand, except for the ND6 and eight tRNA genes that were transcribed from the L-strand. Eleven of the 13 PCGs were used ATG as a start codon, except for ND2 and COXI which began with GTG. Furthermore, there are five types of stop codons, including TAA for ND2, COXII, ATPase6, ATPase8, ND4L, Cytb; AGA for ND5, ND1; TAG for ND6; T- for ND4, COXIII; TA- for ND3. Similar to previous studies, two ribosomal RNA genes were located between the tRNAPhe and tRNALeu which separated by the tRNAVal. We also found that the two control regions were located between the tRNAThr and tRNAPhe which was separated by the tRNAPro, ND6, and tRNAGlu (Huang et al. Citation2015).
The phylogenetic tree illustrated that nine species were mainly divided into two clades, among which the genus Yuhina and the Zosterops were clustered together, and other species were clustered together (). Additionally, the Black-chinned Yuhina was the nearest sister to Zosterops which was consistent with the previous studies (Zhang et al. Citation2007; Cibois et al. Citation2002). Therefore, our work provides molecular data for the taxonomy of Black-chinned Yuhina.
Acknowledgements
We thank the Qian Su, Xue Liu, Pu Zhao, Yuhan Wu, Tianrui Xia, Cong Wang, Kechu Zhang for their assistance in experiments and data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- BirdLife International. 2016. Yuhina nigrimenta. The IUCN Red List of Threatened Species 2016: e. T22716750A94509276. Downloaded on 04 October 2018.
- Cheng T-h. 1987. A synopsis of the avifauna of China. Parey Scientific Publ.
- Cibois A, Kalyakin MV, Lian-Xian H, Pasquet E. 2002. Molecular phylogenetics of babblers (Timaliidae): revaluation of the genera Yuhina and Stachyris. J Avian Biol. 33:380–390.
- Gelang M, Cibois A, Pasquet E, Olsson U, Alström P, Ericson PGP. 2009. Phylogeny of babblers (Aves, Passeriformes): major lineages, family limits and classification. Zoologica Scripta. 38:225–236.
- Gill F, Donsker D, editors. 2018. IOC World Bird List (v8.2).
- Huang R, Zhou Y, Yao Y, Zh B, Zhang Y, Xu H. 2015. Complete mitochondrial genome and phylogenetic relationship analysis of Garrulax affinis (Passeriformes, Timaliidae). Mitochondrial DNA. 27:3502–3503.
- Li B, Yao Y, Li D, Ni Q, Zhang M, Xie M, Xu H. 2017. The complete mitochondrial genome sequence of White-collared Yuhina. Mitochondrial DNA Part A. 28:21–22.
- Moyle RG, Andersen MJ, Oliveros CH, Steinheimer FD, Reddy S. 2012. Phylogeny and biogeography of the core babblers (Aves: Timaliidae). Syst Biol. 61:631–651.
- Sambrock J, Russel DW. 2001. Molecular cloning: a laboratory manual (3rd edition). Immunology. 49:895–909.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Zhang S, Yang L, Yang X, Yang J. 2007. Molecular phylogeny of the Yuhinas (Sylviidae: Yuhina): a paraphyletic group of babblers including Zosterops and Philippine Stachyris. J Ornithol. 148:417–426.
- Zhou Y, Qi Y, Xu H, Huan Z, Li D, Xie M, Ni Q, Zhang M, Yao Y. 2016. The complete mitochondrial genome sequence of Garrulax ocellatus (Aves, Passeriformes, Timaliidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2689–2690.