Abstract
The complete mitochondrial genome of Notonecta montandoni Motschulsky (Hemiptera: Notonectidae) was sequenced in this study. The whole length of the mt genome is 15,150 bp. Similar to other Notonectidae species, it contains a typically conserved structure including 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and a 557 bp long control region. All protein-coding genes started with ATN codons except for COI (TTG) and ended with TAA or TAG except for COII (T), COIII (T), ATP6 (T), and ND3 (T). The complete mitochondrial genome sequence provided here would be useful for further understanding the evolution of backswimmers.
Notonecta montandoni Motschulsky (Hemiptera, Notonectidae) backstroke in the pool with oar-like legs. Notonectids are the most common bugs preying upon both mosquito larvae and adults, an important factor in reducing the mosquito population and considered promising in mosquito control (Shaalan and Canyon Citation2009; Fischer et al. Citation2012). In this study, the complete mitochondrial genome of N. montandoni from Sandaogang Village (N33°39′13″, E111°38′3″), Xixia County, Nanyang City, Henan Province, China, on 18 July 2018 was newly analyzed. The sample of captive N. montandoni was deposited in the Entomological Specimen Room of Taiyuan Normal University, Taiyuan, China.
The complete mt genome, assembled from Illumina sequencing data and submitted to GenBank (accession: MH780861), was a 15,150 bp, a typical double-stranded circular molecule with an asymmetric nucleotide composition (33.12% A, 10.53% C, 13.74% G, and 42.61% T). The order and orientation of 37 genes (including 13 PCGs, 22 tRNAs, and 2 rRNAs) were identical to those of most Notonectidae species (Hua et al. Citation2009). Of these, 14 (trnQ-Gln, trnC-Cys, trnY-Tyr, trnF-Phe, ND5, trnH-His, ND4, ND4L, trnP-Pro, ND1, trnL-Leu (CUN), 16S rRNA, trnV-Val, and 12S rRNA) were located on the minor coding strand (N-strand); however, the other 22 genes and the D-loop region were on the major coding strand (J-strand).
All PCGS shared the start codon ATN (ATA, n = 4; ATT, n = 1; ATG, n = 7), except COI which started with TTG. Seven PCGs terminated with TAA, the COII, COIII, ATP6, and ND3 genes with T, and the ND4 and Cytb genes with TAG. The 22 tRNAs ranged from 62 to 72 bp, and all had a typical cloverleaf secondary structure except trnS-Ser (GUC) which lacked a dihydrouridine arm. The two rRNAs were 766 bp (12S rRNA) and 1018 bp (16S rRNA) long and were separated from each other by trnV-Val. The D-loop region was located between 12S rRNA and trnI-Ile with a total length of 557 bp.
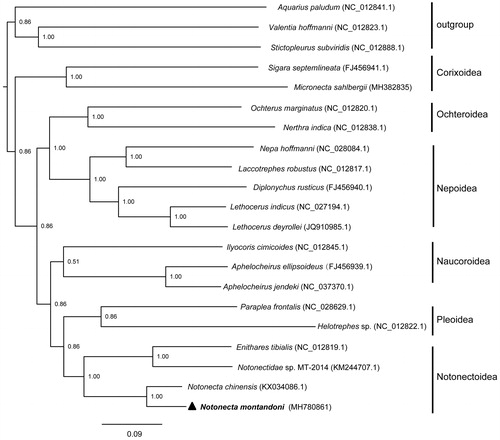
Phylogenetic analysis was constructed by applying 13 PCGs with 18 Notonectidae taxa and three outgroups (). MrBayes v3.2.3 (Ronquist et al. Citation2012) was employed to reconstruct the phylogenetic tree with the GTR + I + G model estimated by Modeltest v 3.7 (Posada and Crandall Citation1998). The phylogram strongly indicated that Notonectoidea was a sister group to Pleoidea (Hebsgaard et al. Citation2004; Li et al. Citation2014). In Notonectoidea, our analysis supported that Enithares and Notonecta was a monophyletic group supported by a posterior probability of 1.00. The complete sequence of the N. montandoni mitochondrial genome provides additional genetic tools for studying population genetics, speciation and biogeography of this group.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Fischer S, Pereyra D, Fernández L. 2012. Predation ability and non-consumptive effects of Notonecta sellata (Heteroptera: Notonectidae) on immature stages of Culex pipiens (Diptera: Culicidae). J Vector Ecol. 37:245–251.
- Hebsgaard MB, Andersen NM, Jakob D. 2004. Phylogeny of the true water bugs (Nepomorpha: Hemiptera–Heteroptera) based on 16S and 28S rDNA and morphology. Syst Entomol. 29:488–508.
- Hua JM, Li M, Dong PZ, Cui Y, Xie Q, Bu WJ. 2009. Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): evidence from mitochondrial genomes. BMC Evol Biol. 9:134.
- Li T, Hua JM, Wright AM, Cui Y, Xie Q, Bu WJ, Hillis DM. 2014. Long-branch attraction and the phylogeny of true water bugs (Hemiptera: Nepomorpha) as estimated from mitochondrial genomes. BMC Evol Biol. 14:1–12.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Shaalan EA, Canyon DV. 2009. Aquatic insect predators and mosquito control. Trop Biomed. 26:223–261.